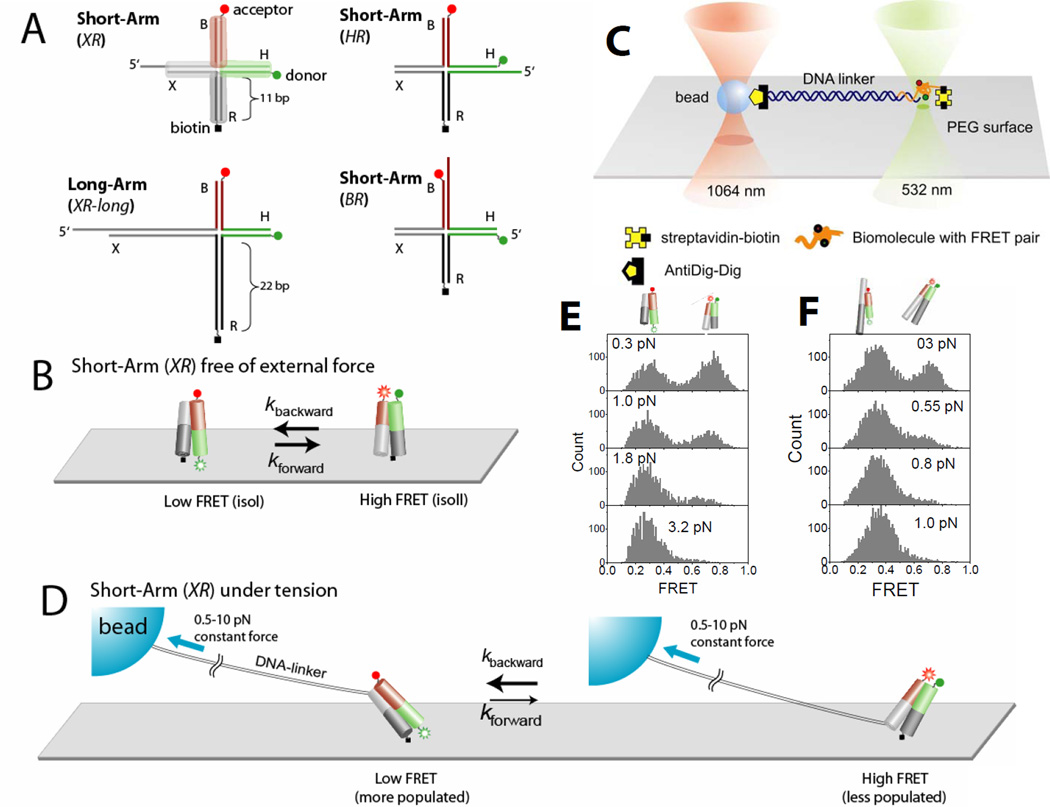
Fig. 1.
Holliday junction constructs and experimental scheme. (A) The HJ species studied. Junction XR comprises four arms of 11 bp, termed B (red), H (green), R (dark gray) and X (gray). Cy3 and Cy5 fluorophores are terminally attached to H and B arms respectively, and the molecule is tethered to the surface through biotin attached to the end of the R arm. Stretching force is applied through the λ-DNA linker hybridized to the X arm. In junction XR-long the lengths of arms R and X are increased to 21 bp. In junction HR the λ-DNA linker is hybridized to the H arm. In junctions HR and BR the λ-DNA linker is hybridized to the H and B arms respectively. (B) Junction XR is known to alternate between two different stacking conformers, isoI (Low FRET) and isoII (High FRET) with similar populations in both states. (C) A surface-immobilized biomolecule with FRET labeling is connected to a trapped bead via a long DNA linker. The linker DNA spatially separates the confocal beam (532 nm) from the trapping beam (1064 nm) such that enhanced photobleaching and an overwhelming background signal induced by the intense trapping laser are avoided. To apply force, the surface immobilized molecule was moved relative to the trapped bead. The confocal beam was programmed to follow the motion of the molecule using the mapping generated between sample scanning and beam scanning (Fig. S6). (D) Force is expected to bias the junction XR to isoI which possesses a larger separation between the two tether points than isoII. (E) FRET histograms of a single junction XR as a function of force. (F) FRET histograms of a single junction XR-long as a function of force.