Abstract
AIM: To develop and validate a case definition of eosinophilic esophagitis (EoE) in the linked Danish health registries.
METHODS: For case definition development, we queried the Danish medical registries from 2006-2007 to identify candidate cases of EoE in Northern Denmark. All International Classification of Diseases-10 (ICD-10) and prescription codes were obtained, and archived pathology slides were obtained and re-reviewed to determine case status. We used an iterative process to select inclusion/exclusion codes, refine the case definition, and optimize sensitivity and specificity. We then re-queried the registries from 2008-2009 to yield a validation set. The case definition algorithm was applied, and sensitivity and specificity were calculated.
RESULTS: Of the 51 and 49 candidate cases identified in both the development and validation sets, 21 and 24 had EoE, respectively. Characteristics of EoE cases in the development set [mean age 35 years; 76% male; 86% dysphagia; 103 eosinophils per high-power field (eos/hpf)] were similar to those in the validation set (mean age 42 years; 83% male; 67% dysphagia; 77 eos/hpf). Re-review of archived slides confirmed that the pathology coding for esophageal eosinophilia was correct in greater than 90% of cases. Two registry-based case algorithms based on pathology, ICD-10, and pharmacy codes were successfully generated in the development set, one that was sensitive (90%) and one that was specific (97%). When these algorithms were applied to the validation set, they remained sensitive (88%) and specific (96%).
CONCLUSION: Two registry-based definitions, one highly sensitive and one highly specific, were developed and validated for the linked Danish national health databases, making future population-based studies feasible.
Keywords: Eosinophilic esophagitis, Denmark, Epidemiology, Case definition, Sensitivity, Specificity
INTRODUCTION
Eosinophilic esophagitis (EoE) is a chronic immune-mediated disorder characterized by symptoms of esophageal dysfunction and infiltration of the esophageal mucosa by eosinophils[1]. While the first case was described in 1978[2], a set of case series in the early 1990s highlighted what are now viewed as the classic clinicopathologic features of EoE[3-5]. Since that time, there has been a remarkable increase in the incidence and prevalence of EoE[6-14], EoE has been detected in up to 16% of patients undergoing upper endoscopy for dysphagia[15-17], and EoE is now the leading cause of food impaction in patients presenting to emergency departments[18-20]. Most data regarding the evolving epidemiology of EoE originate from tertiary centers[7-13], and there have been few population-based studies of EoE across centers or at a national level.
The national population-based medical registries of Denmark, which link medical records, diagnostic codes, pathology findings, and prescription medications via an individual identifier assigned to every person in the country[21-24], offer a unique opportunity to systematically study the epidemiology of EoE. However, no validated registry-based case definitions of EoE exist, and this limitation hampers any attempt to accurately study EoE in Denmark and other countries where national health databases are maintained.
The aims of this study were to characterize candidate and definite EoE cases in Northern Denmark detected through a search of the health registries, and to develop and validate a registry-based case definition of EoE in the linked Danish clinical, pathology, and pharmacy databases that could be used for future epidemiologic study.
MATERIALS AND METHODS
Denmark health registries
This retrospective study of the Danish medical registries was approved by both the University of North Carolina IRB and the Danish Data Protection Agency (record number 2010-41-4986). Denmark, with its stable population of approximately 5.5 million people, is well-suited for epidemiological studies[21-24]. The establishment of the Civil Registration System in 1968 allows information about the same person to be linked across independent registries by using the Civilian Registration Number, a unique identifier assigned to every person in the country[23].
Three of the comprehensive national medical registries in Denmark were utilized to generate the necessary data for this study. These included: the Danish National Registry of Patients, which houses International Classification of Diseases-10 (ICD-10) codes dating from 1994, hospital admission and discharge codes, surgical procedure codes, and outpatient visit codes[25]; the National Pathology Registry, which contains Systematized Nomenclature of Medicine (SNOMED) codes for pathologic specimens dating from 1997[24]; and the Aarhus University Prescription Database, which has outpatient prescription data for Northern Denmark using Anatomical Therapeutic Chemical (ATC) classification system codes[26].
For the purposes of this study we limited our analysis to Northern Denmark. This geographic area contains 1.8 million inhabitants, approximately 1/3 of the Danish population, and allowed ready access to the required pathology specimens for case verification, as described below.
Case definition development
This study utilized two independent sets of patients, one set in which to develop the case definition and one set in which to validate it.
For case definition development, we queried the National Pathology Registry from 2006-2007 in Northern Denmark to identify candidate cases of EoE. These were patients with esophageal eosinophilia as defined by the combination of SNOMED codes for esophageal biopsies (T62xxx) and tissue eosinophilia (M47150). These codes were assigned by the clinical pathologist at the time of specimen examination and interpretation. Next, all ICD-10 diagnostic and ATC prescription codes for these patients were obtained from the National Registry of Patients and the Aarhus University Prescription Database, respectively. Finally, the original archived pathology slides were obtained and, using a previously validated protocol[27], were re-reviewed by the study pathologist (Vyberg M) to determine the maximum eosinophil count [eosinophils per high-power field (eos/hpf); hpf = 0.24 mm2].
From this pool of subjects with esophageal eosinophilia, we identified the subset of patients with a confirmed diagnosis of EoE as per consensus diagnostic guidelines[1,28]. Specifically, these patients had symptoms of esophageal dysfunction, a maximum eosinophil count ≥ 15 eos/hpf on pathology re-review, and no other competing causes of esophageal eosinophilia identified. While the current consensus guidelines allow reflux and EoE to overlap[1], we attempted to minimize this overlap by ensuring that there was not a mixed inflammatory infiltrate on the re-reviewed esophageal biopsies and by assessing use of anti-acid mediations in the months preceding the diagnostic endoscopy (see below). These subjects with confirmed EoE comprised the reference standard for the case definition development set.
We used an iterative process to develop a case definition of EoE based on previously described methodology for gastrointestinal disorders used in the Danish and other administrative databases[29-31]. First, we empirically selected a combination of SNOMED, ICD-10 and ATC codes that were potentially pertinent to the diagnosis of EoE and could exclude disorders on the differential diagnosis for esophageal eosinophilia as well as patients with non-EoE conditions with similar symptom profiles. Next, the National Pathology Registry, National Registry of Patients, and Aarhus University Prescription Database were queried using this combination of codes to provide a set of possible EoE patients. Then, using the confirmed EoE cases previously identified as the reference standard, the sensitivity and specificity of this initial search strategy were determined within the population of patients with esophageal eosinophilia. Sensitivity was calculated by dividing the number of true positive EoE cases identified using the case definition by the total number of reference standard EoE cases. Specificity was calculated by dividing the number of true negative subjects identified using the case definition by the total number of subjects without EoE. By examining the coding and patient characteristics of those classified as false positive and false negative, the administration case definition was further refined, the databases were re-queried with the updated definition, and new operating characteristics were calculated. This process continued until sensitivity and specificity were optimized.
Case definition validation
Using the same methodology as described for case definition development, the case definition was validated in an independent population in Northern Denmark from 2008-2009. In brief, candidate cases of EoE were identified based on SNOMED codes, and a reference standard set of patients with confirmed EoE in the new time frame was created after re-review of original pathology slides and all coding data. The administrative case definition algorithm was then applied to the National Pathology Registry, National Registry of Patients, and Aarhus University Prescription Database, the set of possible EoE cases was generated, and sensitivity and specificity were calculated using the confirmed EoE cases as the reference standard.
RESULTS
Characteristics of the development set
There were 51 patients with esophageal eosinophilia identified in Northern Denmark from 2006-2007, and 21 (41%) had a confirmed diagnosis of EoE (Table 1). This group comprised the reference standard for the development set. Patients with esophageal eosinophilia had a mean age of 42 years, 75% were male, and almost half had dysphagia. While the EoE patients had a similar proportion of males (76%), they were somewhat younger (35 years), and 86% had dysphagia. The maximum eosinophil count was 71 eos/hpf in all patients with esophageal eosinophilia, and 103 eos/hpf in patients with confirmed EoE. The pathology re-review of slides confirmed that the SNOMED coding for esophageal eosinophilia was correct in the vast majority of cases, with 90% meeting criteria for histologic EoE (≥ 15 eos/hpf) (Figure 1).
Table 1.
Characteristics of the case definition development set (2006-2007) and validation set (2008-2009) n (%)
|
Development set (2006-2007) |
Validation set (2007-2008) |
|||||||
| Overall (n = 51) | EoE cases (n = 21) | Sensitive definition (n = 31) | Specific definition (n = 9) | Overall (n = 49) | EoE cases (n = 24) | Sensitive definition (n = 26) | Specific definition (n = 6) | |
| Age, yr (mean ± SD) | 42 ± 19 | 35 ± 16 | 40 ± 18 | 38 ± 19 | 48 ± 21 | 42 ± 15 | 49 ± 19 | 49 ± 19 |
| Male | 38 (75) | 16 (76) | 22 (71) | 7 (78) | 36 (73) | 20 (83) | 21 (81) | 4 (67) |
| Dysphagia | 24 (47) | 18 (86) | 28 (90) | 8 (89) | 19 (39) | 16 (67) | 18 (69) | 3 (50) |
| Esophageal foreign body | 15 (29) | 11 (52) | 14 (45) | 3 (33) | 8 (16) | 2 (8) | 4 (15) | 1 (17) |
| Rhinitis/sinusitis | 5 (10) | 2 (10) | 2 (6) | 1 (11) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Asthma | 3 (6) | 1 (5) | 1 (3) | 0 (0) | 2 (4) | 2 (8) | 2 (8) | 1 (17) |
| PPI use | 38 (75) | 17 (81) | 21 (68) | 9 (100) | 33 (67) | 12 (50) | 21 (81) | 6 (100) |
| Maximum eosinophil count, eos/hpf (mean ± SD) | 71 ± 53 | 103 ± 54 | 84 ± 58 | 93 ± 83 | 68 ± 71 | 77 ± 62 | 60 ± 35 | 43 ± 31 |
EoE: Eosinophilic esophagitis; PPI: Proton-pump inhibitor; eos/hpf: Eosinophils per high-power field.
Figure 1.
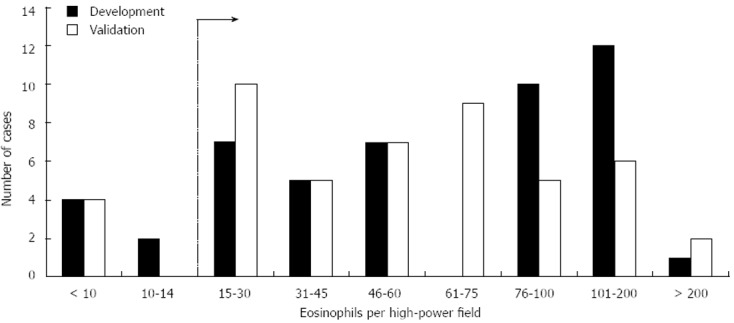
Distribution of levels of esophageal eosinophilia in the development and validation sets of candidate cases of eosinophilic esophagitis identified in the Denmark medical registries. Ranges of esophageal eosinophilia [in eosinophils per high-power field (eos/hpf)] determined after pathology re-review of archived slides are on the x axis. The typical histologic threshold for considering a diagnosis of eosinophilic esophagitis in the appropriate clinical context of 15 eos/hpf is indicated with a dotted line.
Characteristics of the validation set
There were 49 patients with esophageal eosinophilia identified in Northern Denmark from 2008-2009, and 24 (49%) had a confirmed diagnosis of EoE (Table 1). This group comprised the reference standard for the validation set. Patients with esophageal eosinophilia had a mean age of 48 years, 73% were male, and 39% had dysphagia. While the EoE patients had a similar proportion of males (83%), they were also somewhat younger (42 years) and 67% had dysphagia. The maximum eosinophil count was 68 eos/hpf in all patients with esophageal eosinophilia, and 77 eos/hpf in patients with confirmed EoE. Overall, the clinical characteristics of the EoE cases in the development and validation sets were similar, and pathology re-review in the validation set also confirmed correct SNOMED coding in 90% of cases (Figure 1).
EoE case definition algorithms and operating characteristics
The iterative methodology for case definition development and validation yielded two registry-based case diagnostic algorithms, one that was highly sensitive and one that was highly specific. The highly sensitive case definition maximized the number of cases of EoE that were identified in the database search, while the highly specific definition accepted a lower sensitivity (i.e., not identifying all EoE cases) in exchange for ensuring that all cases were true EoE cases (i.e., minimizing false positives).
The final case definition algorithms are presented in Table 2. The “sensitive” algorithm has three steps. The first uses SNOMED codes to include all patients with esophageal eosinophilia. The second excludes patients if they have any one of eleven ICD-10 codes that could cause non-EoE related esophageal eosinophilia or esophageal injury that could mimic EoE clinically. The third requires patients to have at least one of 13 ICD-10 codes for symptoms of esophageal dysfunction. The “specific” algorithm has an additional step to exclude false positives which requires a documented proton-pump inhibitor (PPI) or histamine-2 receptor antagonist prescription within 2 mo of the esophageal biopsy date.
Table 2.
Case definition algorithm
| For both the sensitive and the specific case definitions: |
| (1) Include all patients with systematized nomenclature of medicine M47150 (finding of tissue eosinophil) and T62xxx (esophageal biopsy) |
| (2) Exclude patients if they have one of the following International Classification of Diseases-10 codes occurring at any time: |
| B20.x - human immunodeficiency virus |
| C15.x - malignant neoplasm of esophagus |
| C16.0 - malignant neoplasm of cardia |
| C92.x - myeloid leukemia |
| C94.x - other leukemias of specialized cell type |
| C96.x - other and unspecified lymph/heme malignancies |
| K22.0 - achalasia |
| K50.x - Crohn’s disease |
| K52.8 - eosinophilic gastritis/duodenitis |
| T28.1 - burn of esophagus |
| T28.6 - corrosion of esophagus |
| (3) Include patients if one of the following International Classification of Diseases-10 codes is present at or before the date of the procedure: |
| K22.2 - esophageal obstruction |
| K30.9 - dyspepsia |
| P92.x - feeding problems of the newborn |
| R07.x - pain in throat and chest |
| R10.x - abdominal pain |
| R11.x - nausea/vomiting |
| R12.x - heartburn |
| R13.x - dysphagia |
| T18.0 - foreign body in mouth |
| T18.1 - foreign body in esophagus |
| T18.2 - foreign body in stomach |
| T18.9 - foreign body in genitourinary tract unspecified |
| T98.0 - sequelae of foreign body entering through a natural orifice |
| Additional requirement for the specific case definition: |
| (4) Include patients if there is a proton-pump inhibitor prescription (A02BCxx) or H2RA prescription (A02BAxx) within 2 mo of esophageal biopsy date, up to and including 2 d prior to the esophagogastroduodenoscopy |
When these algorithms were applied to the development set, the clinical characteristics of the cases identified for both the sensitive definition (n = 31; mean age 40 years; 71% male; 90% dysphagia; 84 eos/hpf) and the specific definition (n = 8; mean age 38 years; 78% male; 89% dysphagia; 93 eos/hpf) were similar to those for the reference standard EoE cases (Table 1).
When these algorithms were applied to the validation set, the clinical features for the sensitive definition (n = 26; mean age 49 years; 81% male; 69% dysphagia; 60 eos/hpf) and specific definition (n = 6; mean age 49 years; 67% male; 50% dysphagia; 43 eos/hpf) were also similar to those for the reference standard for EoE cases (Table 1).
In the development set, the “sensitive” algorithm had a sensitivity and specificity of 90% and 60%, and the “specific” algorithm had values of 38% and 97%, respectively (Table 3). When these algorithms were applied to the validation set, the operating characteristics were essentially unchanged, with a sensitivity and specificity of 88% and 80%, for the “sensitive” algorithm, and 21% and 96% for the “specific” algorithms.
Table 3.
Sensitivity and specificity of the case definitions1 n (%)
|
Development set |
Validation set |
|||
| Sensitive definition | Specific definition | Sensitive definition | Specific definition | |
| Sensitivity | 90 (90) | 38 (38) | 88 (88) | 21 (21) |
| Specificity | 60 (60) | 97 (97) | 80 (80) | 96 (96) |
1Sensitivity: Number of true positive eosinophilic esophagitis (EoE) cases identified by the case definition divided by the total number of reference standard EoE cases; Specificity: Number of true negative subjects identified by the case definition divided by the total number of subjects without EoE.
DISCUSSION
The epidemiology of EoE has been rapidly evolving over the past two decades, with a marked increase in incidence and prevalence. The study of EoE in large national or administrative databases, however, has been hampered by the lack of a validated case definition of EoE and the difficulty of developing such a definition due to the clinicopathologic nature of the disorder. This study takes advantage of the fact that the health registries in Denmark can link pathologic data to diagnostic and prescription coding data, thus providing all of the needed components to identify cases of EoE.
There are two main results for this study. The first is that the confirmed EoE cases identified in Denmark via the health registries had similar features to those reported both in Denmark as well as elsewhere in the world[1,7-17,32-35]. The second is that two registry-based case definitions, one highly sensitive and one highly specific, were successfully developed and validated for use in the Danish national health databases. This is the first study to do so, and this result makes future large-scale population-based studies feasible in that country. This is important because the majority of investigations studying the epidemiology of EoE are either from selected counties within a country or region, from single centers but are not population-based, or from larger databases that have limitations.
For example, in the two counties studied in northern Sweden during the Kalixanda study, Ronkainen et al[36] found that 11 of 1000 (1.1%) subjects had prevalent esophageal eosinophilia ≥ 15 eos/hpf. While this is the only published study with a true population-based sampling strategy, these individuals would not necessarily meet current EoE diagnostic guidelines because not all were symptomatic and competing causes of esophageal eosinophilia were not excluded[1,28]. In Olten County, Switzerland, a well-defined geographic region with a stable population and practitioners who are expert in EoE diagnosis, Hruz et al[14] have recently updated their estimates of the incidence (7/100 000) and prevalence (43/100 000) of EoE[12]. Prasad et al[9] reported similar estimates from Olmstead County, Minnesota (incidence 9/100 000; prevalence 55/100 000) in patients identified retrospectively, and Spergel et al[37] derived a similar prevalence estimate (52/100 000) from physician surveys. In all of these studies, however, the included patients and providers were not sampled in population-based frames. Additional retrospective single center studies, while providing important data, are subject to similar limitations[7,8,10,11,13,38,39]. Three prospective studies examining the prevalence of EoE in patients undergoing upper endoscopy for any symptom[17], or for dysphagia provide equally important data[15,16], but are also from single centers and enrolled patients who were actively undergoing evaluation and so cannot necessarily be generalized to an entire population. In a series of studies that utilized a large pathology database in the United States, the national distribution of esophageal eosinophilia and EoE has been confirmed, but the data are also not population based[40-42]. Most recently, abstract data from the national health system in the Netherlands has been presented showing a rapid rise in the incidence of esophageal eosinophilia, but this study did not employ a validated case definition of EoE[43]. The current study attempts to address the challenges encountered in the definition of EoE when using county- or national-level database or registry data, and to set the stage for conducting true population-level EoE research.
There are both limitations and strengths of this study that should be acknowledged. The first issue is whether there could be misclassification of cases of EoE in the reference standard groups. This appears to be unlikely given that histology, symptom coding, and prescription data were used to apply EoE consensus diagnostic guidelines to the study population. Nevertheless, because this is a retrospective analysis, we cannot completely exclude the possibility of overlap between gastroesophageal reflux disease and EoE, and are unable to fully address the issue of PPI-responsive esophageal eosinophilia (PPI-REE). It is interesting to observe, however that only 41% and 49% of subjects with esophageal eosinophilia were confirmed to have EoE in the development and validation sets, respectively, suggesting that half or more of subjects with esophageal eosinophilia do not have EoE. The proportion of subjects with esophageal eosinophilia who did not have EoE in our study is similar to the reported proportion of subjects with esophageal eosinophilia who have PPI-REE, ranging in various studies from approximately 30%-75%[44-48]. The poor specificity of the presence of esophageal eosinophilia for the diagnosis of EoE, coupled with the high proportion of subjects with esophageal eosinophilia who have PPI-REE, emphasize that esophageal eosinophilia alone is not adequate for case-finding of EoE in pathology databases, and cannot be used in isolation to diagnose EoE[1,28,49,50]. A related point is that while it is a strength of this study to have obtained and re-reviewed the original pathology slides, it is important to acknowledge that the biopsies were originally taken for clinical purposes at the discretion of individual endoscopists, and the location and number of biopsies could vary.
It is a strength of the study that two case definitions, one sensitive and one specific, were developed. In instances where overlap or misclassification may be a concern, the “specific” algorithm could be used to ensure the most homogenous EoE population is generated. The two case definitions also allow for bounding and performing sensitivity analyses around epidemiologic estimates. Moreover, the use of the linked Denmark databases is another strength of this study, as there are relatively few data sources that are as rich and contain patient-level pathology information linked to administrative coding. This is illustrated by our ability to obtain and re-review the original glass pathology slides in order to validate the SNOMED codes for esophageal eosinophilia.
The case definitions developed in this study are derived from the Denmark databases and it is unknown whether these would be valid in other settings. However, the methodology presented here could be readily replicated in different databases to validate EoE case definitions, and our proposed algorithm could be used as a starting point in an iterative process to develop case definitions in other databases.
In conclusion, this study of the linked Danish national health registries successfully identified and confirmed cases of EoE in Denmark. Two administrative registry-based case definitions, one highly sensitive to maximize the number of cases of EoE identified, and one highly specific to minimize the number of false positive EoE cases included, were developed and validated. The operating characteristics of the case algorithms are sufficient to support future population-based studies of the epidemiology of EoE in Denmark, and may serve as a template for developing similar definitions in other databases.
COMMENTS
Background
Population-based data on eosinophilic esophagitis (EoE) are lacking. The national medical registries in Denmark offer a unique opportunity to systematically study EoE because these resources link pathology, clinical, and pharmacological data at the patient level. However, no validated registry-based case definitions of EoE exist, and these must be developed prior to conducting large-scale studies.
Research frontiers
EoE is a newly emerging chronic immune-mediated disorder characterized by symptoms of esophageal dysfunction and infiltration of the esophageal mucosa by eosinophils. The epidemiology of EoE is increasingly understood, but much of the data are from single referral centers, and there are few population-based studies published.
Innovations and breakthroughs
This is the first study to systematically develop and validate an administrative case definition of EoE in the Danish databases. The authors present two registry-based definitions, one highly sensitive and one highly specific for EoE.
Applications
These definitions can now be utilized in future population-based studies of EoE in Denmark.
Peer review
This is a well designed study about development and validation of a registry based case definition of EoE in Northern Denmark. It has to be pointed out that in the re-reviewed slides of pathology represents an advantage in methodology.
Footnotes
Supported by Pilot/feasibility Grant from the UNC Center for Gastrointestinal Biology and Disease, NIH P30 DK34987; and NIH award K23DK090073 (in part)
P- Reviewer von Arnim U S- Editor Wen LL L- Editor A E- Editor Xiong L
References
- 1.Liacouras CA, Furuta GT, Hirano I, Atkins D, Attwood SE, Bonis PA, Burks AW, Chehade M, Collins MH, Dellon ES, et al. Eosinophilic esophagitis: updated consensus recommendations for children and adults. J Allergy Clin Immunol. 2011;128:3–20.e6; quiz 21-22. doi: 10.1016/j.jaci.2011.02.040. [DOI] [PubMed] [Google Scholar]
- 2.Landres RT, Kuster GG, Strum WB. Eosinophilic esophagitis in a patient with vigorous achalasia. Gastroenterology. 1978;74:1298–1301. [PubMed] [Google Scholar]
- 3.Attwood SE, Smyrk TC, Demeester TR, Jones JB. Esophageal eosinophilia with dysphagia. A distinct clinicopathologic syndrome. Dig Dis Sci. 1993;38:109–116. doi: 10.1007/BF01296781. [DOI] [PubMed] [Google Scholar]
- 4.Straumann A, Spichtin HP, Bernoulli R, Loosli J, Vögtlin J. [Idiopathic eosinophilic esophagitis: a frequently overlooked disease with typical clinical aspects and discrete endoscopic findings] Schweiz Med Wochenschr. 1994;124:1419–1429. [PubMed] [Google Scholar]
- 5.Kelly KJ, Lazenby AJ, Rowe PC, Yardley JH, Perman JA, Sampson HA. Eosinophilic esophagitis attributed to gastroesophageal reflux: improvement with an amino acid-based formula. Gastroenterology. 1995;109:1503–1512. doi: 10.1016/0016-5085(95)90637-1. [DOI] [PubMed] [Google Scholar]
- 6.Sealock RJ, Rendon G, El-Serag HB. Systematic review: the epidemiology of eosinophilic oesophagitis in adults. Aliment Pharmacol Ther. 2010;32:712–719. doi: 10.1111/j.1365-2036.2010.04411.x. [DOI] [PubMed] [Google Scholar]
- 7.Spergel JM, Brown-Whitehorn TF, Beausoleil JL, Franciosi J, Shuker M, Verma R, Liacouras CA. 14 years of eosinophilic esophagitis: clinical features and prognosis. J Pediatr Gastroenterol Nutr. 2009;48:30–36. doi: 10.1097/MPG.0b013e3181788282. [DOI] [PubMed] [Google Scholar]
- 8.Dellon ES, Gibbs WB, Fritchie KJ, Rubinas TC, Wilson LA, Woosley JT, Shaheen NJ. Clinical, endoscopic, and histologic findings distinguish eosinophilic esophagitis from gastroesophageal reflux disease. Clin Gastroenterol Hepatol. 2009;7:1305–1313; quiz 1261. doi: 10.1016/j.cgh.2009.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prasad GA, Alexander JA, Schleck CD, Zinsmeister AR, Smyrk TC, Elias RM, Locke GR, Talley NJ. Epidemiology of eosinophilic esophagitis over three decades in Olmsted County, Minnesota. Clin Gastroenterol Hepatol. 2009;7:1055–1061. doi: 10.1016/j.cgh.2009.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gill R, Durst P, Rewalt M, Elitsur Y. Eosinophilic esophagitis disease in children from West Virginia: a review of the last decade (1995-2004) Am J Gastroenterol. 2007;102:2281–2285. doi: 10.1111/j.1572-0241.2007.01352.x. [DOI] [PubMed] [Google Scholar]
- 11.Noel RJ, Putnam PE, Rothenberg ME. Eosinophilic esophagitis. N Engl J Med. 2004;351:940–941. doi: 10.1056/NEJM200408263510924. [DOI] [PubMed] [Google Scholar]
- 12.Straumann A, Simon HU. Eosinophilic esophagitis: escalating epidemiology? J Allergy Clin Immunol. 2005;115:418–419. doi: 10.1016/j.jaci.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 13.Cherian S, Smith NM, Forbes DA. Rapidly increasing prevalence of eosinophilic oesophagitis in Western Australia. Arch Dis Child. 2006;91:1000–1004. doi: 10.1136/adc.2006.100974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hruz P, Straumann A, Bussmann C, Heer P, Simon HU, Zwahlen M, Beglinger C, Schoepfer AM. Escalating incidence of eosinophilic esophagitis: a 20-year prospective, population-based study in Olten County, Switzerland. J Allergy Clin Immunol. 2011;128:1349–1350.e5. doi: 10.1016/j.jaci.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 15.Prasad GA, Talley NJ, Romero Y, Arora AS, Kryzer LA, Smyrk TC, Alexander JA. Prevalence and predictive factors of eosinophilic esophagitis in patients presenting with dysphagia: a prospective study. Am J Gastroenterol. 2007;102:2627–2632. doi: 10.1111/j.1572-0241.2007.01512.x. [DOI] [PubMed] [Google Scholar]
- 16.Mackenzie SH, Go M, Chadwick B, Thomas K, Fang J, Kuwada S, Lamphier S, Hilden K, Peterson K. Eosinophilic oesophagitis in patients presenting with dysphagia--a prospective analysis. Aliment Pharmacol Ther. 2008;28:1140–1146. doi: 10.1111/j.1365-2036.2008.03795.x. [DOI] [PubMed] [Google Scholar]
- 17.Veerappan GR, Perry JL, Duncan TJ, Baker TP, Maydonovitch C, Lake JM, Wong RK, Osgard EM. Prevalence of eosinophilic esophagitis in an adult population undergoing upper endoscopy: a prospective study. Clin Gastroenterol Hepatol. 2009;7:420–426, 426.e1-2. doi: 10.1016/j.cgh.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 18.Desai TK, Stecevic V, Chang CH, Goldstein NS, Badizadegan K, Furuta GT. Association of eosinophilic inflammation with esophageal food impaction in adults. Gastrointest Endosc. 2005;61:795–801. doi: 10.1016/s0016-5107(05)00313-5. [DOI] [PubMed] [Google Scholar]
- 19.Kerlin P, Jones D, Remedios M, Campbell C. Prevalence of eosinophilic esophagitis in adults with food bolus obstruction of the esophagus. J Clin Gastroenterol. 2007;41:356–361. doi: 10.1097/01.mcg.0000225590.08825.77. [DOI] [PubMed] [Google Scholar]
- 20.Sperry SL, Crockett SD, Miller CB, Shaheen NJ, Dellon ES. Esophageal foreign-body impactions: epidemiology, time trends, and the impact of the increasing prevalence of eosinophilic esophagitis. Gastrointest Endosc. 2011;74:985–991. doi: 10.1016/j.gie.2011.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frank L. Epidemiology. When an entire country is a cohort. Science. 2000;287:2398–2399. doi: 10.1126/science.287.5462.2398. [DOI] [PubMed] [Google Scholar]
- 22.Frank L. Epidemiology. The epidemiologist’s dream: Denmark. Science. 2003;301:163. doi: 10.1126/science.301.5630.163. [DOI] [PubMed] [Google Scholar]
- 23.Sorensen HT, Christensen T, Schlosser HK, Pedersen L. Use of Medical Databases in Clinical Epidemiology. Aarhus, Denmark: Department of Clinical Epidemiology, Aarhus University Hospital; 2008. [Google Scholar]
- 24.Erichsen R, Lash TL, Hamilton-Dutoit SJ, Bjerregaard B, Vyberg M, Pedersen L. Existing data sources for clinical epidemiology: the Danish National Pathology Registry and Data Bank. Clin Epidemiol. 2010;2:51–56. doi: 10.2147/clep.s9908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andersen TF, Madsen M, Jørgensen J, Mellemkjoer L, Olsen JH. The Danish National Hospital Register. A valuable source of data for modern health sciences. Dan Med Bull. 1999;46:263–268. [PubMed] [Google Scholar]
- 26.Ehrenstein V, Antonsen S, Pedersen L. Existing data sources for clinical epidemiology: Aarhus University Prescription Database. Clin Epidemiol. 2010;2:273–279. doi: 10.2147/CLEP.S13458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dellon ES, Fritchie KJ, Rubinas TC, Woosley JT, Shaheen NJ. Inter- and intraobserver reliability and validation of a new method for determination of eosinophil counts in patients with esophageal eosinophilia. Dig Dis Sci. 2010;55:1940–1949. doi: 10.1007/s10620-009-1005-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Furuta GT, Liacouras CA, Collins MH, Gupta SK, Justinich C, Putnam PE, Bonis P, Hassall E, Straumann A, Rothenberg ME. Eosinophilic esophagitis in children and adults: a systematic review and consensus recommendations for diagnosis and treatment. Gastroenterology. 2007;133:1342–1363. doi: 10.1053/j.gastro.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 29.Fonager K, Sørensen HT, Rasmussen SN, Møller-Petersen J, Vyberg M. Assessment of the diagnoses of Crohn’s disease and ulcerative colitis in a Danish hospital information system. Scand J Gastroenterol. 1996;31:154–159. doi: 10.3109/00365529609031980. [DOI] [PubMed] [Google Scholar]
- 30.Herrinton LJ, Liu L, Lewis JD, Griffin PM, Allison J. Incidence and prevalence of inflammatory bowel disease in a Northern California managed care organization, 1996-2002. Am J Gastroenterol. 2008;103:1998–2006. doi: 10.1111/j.1572-0241.2008.01960.x. [DOI] [PubMed] [Google Scholar]
- 31.Liu L, Allison JE, Herrinton LJ. Validity of computerized diagnoses, procedures, and drugs for inflammatory bowel disease in a northern California managed care organization. Pharmacoepidemiol Drug Saf. 2009;18:1086–1093. doi: 10.1002/pds.1824. [DOI] [PubMed] [Google Scholar]
- 32.Dalby K, Nielsen RG, Kruse-Andersen S, Fenger C, Bindslev-Jensen C, Ljungberg S, Larsen K, Walsted AM, Husby S. Eosinophilic oesophagitis in infants and children in the region of southern Denmark: a prospective study of prevalence and clinical presentation. J Pediatr Gastroenterol Nutr. 2010;51:280–282. doi: 10.1097/MPG.0b013e3181d1b107. [DOI] [PubMed] [Google Scholar]
- 33.Dalby K, Nielsen RG, Kruse-Andersen S, Fenger C, Durup J, Husby S. Gastroesophageal reflux disease and eosinophilic esophagitis in infants and children. A study of esophageal pH, multiple intraluminal impedance and endoscopic ultrasound. Scand J Gastroenterol. 2010;45:1029–1035. doi: 10.3109/00365521.2010.487917. [DOI] [PubMed] [Google Scholar]
- 34.Dellon ES, Aderoju A, Woosley JT, Sandler RS, Shaheen NJ. Variability in diagnostic criteria for eosinophilic esophagitis: a systematic review. Am J Gastroenterol. 2007;102:2300–2313. doi: 10.1111/j.1572-0241.2007.01396.x. [DOI] [PubMed] [Google Scholar]
- 35.Gonsalves N, Policarpio-Nicolas M, Zhang Q, Rao MS, Hirano I. Histopathologic variability and endoscopic correlates in adults with eosinophilic esophagitis. Gastrointest Endosc. 2006;64:313–319. doi: 10.1016/j.gie.2006.04.037. [DOI] [PubMed] [Google Scholar]
- 36.Ronkainen J, Talley NJ, Aro P, Storskrubb T, Johansson SE, Lind T, Bolling-Sternevald E, Vieth M, Stolte M, Walker MM, Agréus L. Prevalence of oesophageal eosinophils and eosinophilic oesophagitis in adults: the population-based Kalixanda study. Gut. 2007;56:615–620. doi: 10.1136/gut.2006.107714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spergel JM, Book WM, Mays E, Song L, Shah SS, Talley NJ, Bonis PA. Variation in prevalence, diagnostic criteria, and initial management options for eosinophilic gastrointestinal diseases in the United States. J Pediatr Gastroenterol Nutr. 2011;52:300–306. doi: 10.1097/MPG.0b013e3181eb5a9f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Franciosi JP, Tam V, Liacouras CA, Spergel JM. A case-control study of sociodemographic and geographic characteristics of 335 children with eosinophilic esophagitis. Clin Gastroenterol Hepatol. 2009;7:415–419. doi: 10.1016/j.cgh.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 39.DeBrosse CW, Collins MH, Buckmeier Butz BK, Allen CL, King EC, Assa’ad AH, Abonia JP, Putnam PE, Rothenberg ME, Franciosi JP. Identification, epidemiology, and chronicity of pediatric esophageal eosinophilia, 1982-1999. J Allergy Clin Immunol. 2010;126:112–119. doi: 10.1016/j.jaci.2010.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kapel RC, Miller JK, Torres C, Aksoy S, Lash R, Katzka DA. Eosinophilic esophagitis: a prevalent disease in the United States that affects all age groups. Gastroenterology. 2008;134:1316–1321. doi: 10.1053/j.gastro.2008.02.016. [DOI] [PubMed] [Google Scholar]
- 41.Dellon ES, Peery AF, Shaheen NJ, Morgan DR, Hurrell JM, Lash RH, Genta RM. Inverse association of esophageal eosinophilia with Helicobacter pylori based on analysis of a US pathology database. Gastroenterology. 2011;141:1586–1592. doi: 10.1053/j.gastro.2011.06.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hurrell JM, Genta RM, Dellon ES. Prevalence of esophageal eosinophilia varies by climate zone in the United States. Am J Gastroenterol. 2012;107:698–706. doi: 10.1038/ajg.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Rhijn BD, Verheij J, Smout AJ, Bredenoord AJ. Rapidly increasing incidence of eosinophilic esophagitis in a large cohort. Neurogastroenterol Motil. 2013;25:47–e5. doi: 10.1111/nmo.12009. [DOI] [PubMed] [Google Scholar]
- 44.Molina-Infante J, Ferrando-Lamana L, Ripoll C, Hernandez-Alonso M, Mateos JM, Fernandez-Bermejo M, Dueñas C, Fernandez-Gonzalez N, Quintana EM, Gonzalez-Nuñez MA. Esophageal eosinophilic infiltration responds to proton pump inhibition in most adults. Clin Gastroenterol Hepatol. 2011;9:110–117. doi: 10.1016/j.cgh.2010.09.019. [DOI] [PubMed] [Google Scholar]
- 45.Sayej WN, Patel R, Baker RD, Tron E, Baker SS. Treatment with high-dose proton pump inhibitors helps distinguish eosinophilic esophagitis from noneosinophilic esophagitis. J Pediatr Gastroenterol Nutr. 2009;49:393–399. doi: 10.1097/MPG.0b013e31819c4b3e. [DOI] [PubMed] [Google Scholar]
- 46.Dranove JE, Horn DS, Davis MA, Kernek KM, Gupta SK. Predictors of response to proton pump inhibitor therapy among children with significant esophageal eosinophilia. J Pediatr. 2009;154:96–100. doi: 10.1016/j.jpeds.2008.07.042. [DOI] [PubMed] [Google Scholar]
- 47.Peterson KA, Thomas KL, Hilden K, Emerson LL, Wills JC, Fang JC. Comparison of esomeprazole to aerosolized, swallowed fluticasone for eosinophilic esophagitis. Dig Dis Sci. 2010;55:1313–1319. doi: 10.1007/s10620-009-0859-4. [DOI] [PubMed] [Google Scholar]
- 48.Moawad FJ, Dias JA, Veerappan GR, Baker T, Maydonovitch CL. Comparison of aerosolized swallowed fluticasone to esomeprazole for the treatment of eosinophilic esophagitis. Am J Gastroenterol. 2011;106(Suppl 2):S12. doi: 10.1038/ajg.2012.443. [DOI] [PubMed] [Google Scholar]
- 49.Rodrigo S, Abboud G, Oh D, DeMeester SR, Hagen J, Lipham J, DeMeester TR, Chandrasoma P. High intraepithelial eosinophil counts in esophageal squamous epithelium are not specific for eosinophilic esophagitis in adults. Am J Gastroenterol. 2008;103:435–442. doi: 10.1111/j.1572-0241.2007.01594.x. [DOI] [PubMed] [Google Scholar]
- 50.Dellon ES. Diagnosis and management of eosinophilic esophagitis. Clin Gastroenterol Hepatol. 2012;10:1066–1078. doi: 10.1016/j.cgh.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]


