FIGURE 2.
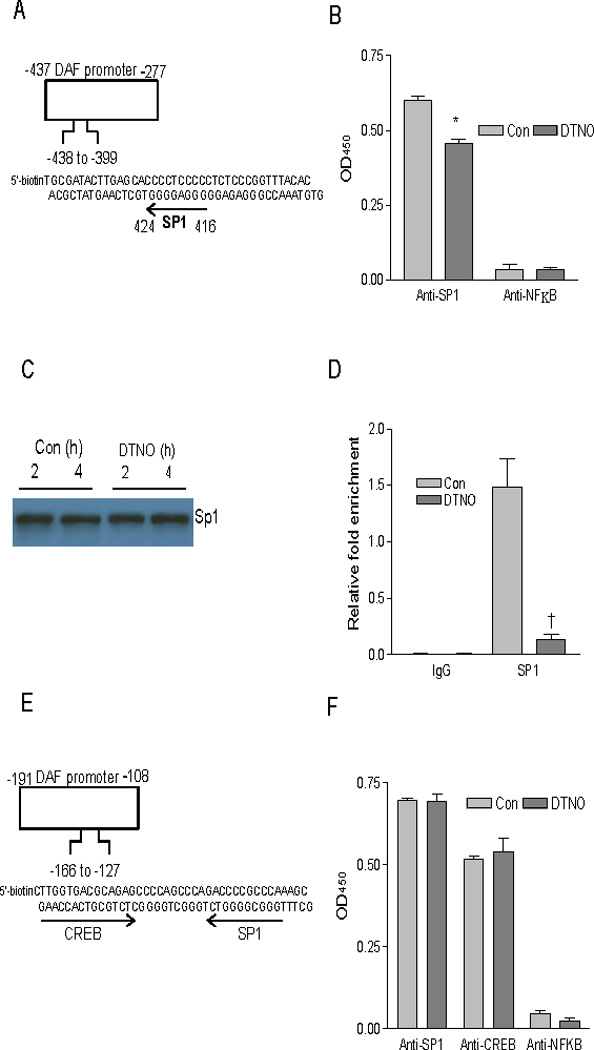
Binding of transcription factor Sp1 to DAF promoter between nucleotides − 424 and − 416 is altered by NO. (A) Nucleotide sequence of 5’-biotin tagged DAF promoter fragment between nucleotides − 438 to − 399 containing one putative Sp1 binding site. (B) Ishikawa cells were treated with DTNO (1 mM) for 4 h. Nuclear fractions from control and DTNO treated cells equivalent to 1.75 µg of protein were added to DAF promoter fragment (−438 to −399) attached to streptavidin coated wells in a micro plate. The amount of Sp1 protein bound to DAF promoter was measured using anti-Sp1 antibody and secondary antibody conjugated to HRP. (*, p < 0.0003 versus control). (C) Expression of Sp1 protein in nuclear extracts of control and DTNO treated cells was assessed by western blotting. (D) ChIP and quantitative qRT-PCR. The control and DTNO treated cells were fixed with formaldehyde, and cross-linked, and the chromatin was sheared. The chromatin was immunoprecipitated with anti-Sp1 antibody or IgG. The binding of Sp1 to the DAF promoter fragments − 438 to − 399 was measured by quantitative real time PCR using specific primers and relative quantitation method. The amounts of immunoprecipitated DNA were normalized to the inputs and plotted (†, p < 0.0063 versus control). (E) Nucleotide sequence of 5’-biotin tagged DAF promoter fragment between nucleotides − 166 to − 127 containing one putative Sp1 binding site and one putative CREB binding site. (F) Nuclear fractions from control and DTNO treated cells equivalent to 1.75 µg of protein were added to DAF promoter fragment (−166 to −127) attached to streptavidin coated wells in a micro plate. The amount of Sp1 or CREB protein bound to DAF promoter was measured using anti-Sp1 or anti-CREB antibody and secondary antibody conjugated to HRP. All experiments were performed three times, and one set of data is presented. Error bars indicate S.E of triplicate samples.
