Abstract
Human cyclin F was originally isolated as a cDNA capable of suppressing the temperature sensitivity of a Saccharomyces cerevisiae cdc4-1 mutant. Its tightly regulated expression and high conservation in the evolutionary progression from amphibians to mammals suggest that it coordinates the timing of a critical cell cycle event. The fact that it contains an F box and can form an SCF (Skp1-Cul1/Cdc53-F-box) complex in vivo further suggests that it may also function in proteolysis. To investigate the role of cyclin F in vivo, we generated mice deficient for cyclin F and conditionally deficient mice as well as mouse embryonic fibroblasts (MEFs) conditionally deficient for cyclin F. Heterozygous animals are normal and fertile, but CycF−/− animals, with a myriad of developmental anomalies due in large part to failures in yolk sac and chorioallantoic placentation, die around embryonic day 10.5. Tissue-specific deletion of cyclin F revealed that it was not required for the development and function of a number of different embryonic and adult tissues. In contrast, MEFs lacking cyclin F, while viable, do exhibit cell cycle defects, including reduced population-doubling time and a delay in cell cycle reentry from quiescence, indicating that cyclin F plays a role in cell cycle regulation.
The successful completion of the cell division cycle relies in large part on the appropriate temporal control of gene expression. The cell cycle is organized as a series of biochemical reactions designed to replicate cellular DNA and distribute it equally to two daughter cells with high fidelity. Cyclins are key components of the core cell cycle machinery. Although cyclins were initially characterized as molecules whose expression “cycled” (hence the name) once per cell cycle, it is now known that they function both as activating regulatory subunits and substrate specificity-determining components for cyclin-dependent kinases (Cdks) (9, 27, 29). It is the ordered periodicity with which they are expressed that dictates the appropriate execution of the cell cycle program.
Loss-of-function studies in mice have proven particularly useful in understanding specific roles carried out by cyclins and the tissues which are exquisitely dependent upon their activity. Additionally, such mouse models have facilitated the generation of cellular reagents to further dissect the biochemical functions carried out by cyclins (3, 4, 10, 13, 21, 23, 32, 33, 39). For example, knockout studies have revealed that each of the D-type cyclins shows tissue-specific defects. This is not due to disparate biochemical capabilities but rather to tissue-specific expression patterns, since each D cyclin appears to be capable of substituting for the other (4, 10, 32, 33). The fact that mice lacking two D cyclins exhibit more exacerbated phenotypes in certain tissues than the mice lacking either cyclin D alone also supports the notion that they have similar biochemical functions (4). In addition, cyclin E expression from the cyclin D1 promoter completely rescues phenotypic consequences of cyclin D1 loss by bypassing cyclin D1's function in the cell cycle (12), suggesting that the essential role of cyclin D is to activate cyclin E expression. Surprisingly, the E-type cyclins appear to be dispensable for cell cycle progression in normally cycling cells, but they are essential for cell cycle reentry (13, 24).
The mitotic A- and B-type cyclins are not formally interchangeable, since animals carrying single mutations of cyclin A2 or cyclin B1 result in embryonic lethality (3, 23). While, within the cyclin A class, cyclins A1 and A2 may have similar biochemical capabilities, they do not exhibit formal redundancy because they are not expressed in the same tissues. Similarly, the inability of cyclin B2 to compensate for cyclin B1's loss reflects their different subcellular localizations, indicating different biochemical functions. Still, if cyclin B2 is altered so that it has cyclin B1's subcellular localization, it then compensates for B1's functions (8). Thus, mouse models have proven particularly useful in furthering our mechanistic understanding of cyclin biology (reviewed in reference 39).
Cyclin F was initially identified as a human cDNA capable of suppressing the temperature sensitivity of a Saccharomyces cerevisiae cdc4-1 mutant (1). cdc4-1 arrests at the nonpermissive temperature at the G1/S boundary with high levels of the Cdk inhibitor Sic1 (31). Cyclin F is thought to bypass this arrest by titrating away Sic1 (2). This facilitates Cdk activation, which drives cell cycle progression. Cyclin F shares the greatest amino acid sequence similarity with cyclin A, both in its cyclin box and beyond. Early characterizations of cyclin F confirmed that, like other cyclins, both its mRNA and protein are expressed in a tightly cell cycle-regulated fashion. Its expression begins in S phase, peaks in G2, and disappears as cells enter mitosis. It also becomes phosphorylated just before cells enter mitosis (1). Subsequent work confirmed that, in a pattern mirroring cyclin A2 and cyclin B1, cyclin F is expressed in all actively dividing cells of both the embryo and the adult (2a). This expression is in contrast to that of CDK inhibitors such as p21, whose expression generally correlates with cell cycle exit (25).
Though they are otherwise functionally dissimilar, cyclin F and Cdc4 were revealed by subsequent analyses to share a motif. This motif has since been discovered in a variety of proteins conserved in the evolutionary progression from yeast to humans and is called the F-box motif (2). F-box proteins function as substrate specificity-determining components of the SCF (Skp1-Cul1/Cdc53-F-box) E3 ubiquitin ligase complex. Polyubiquitination is the rate-limiting step in regulating protein stability. It involves a three-step cascade of ubiquitin transfer reactions. First, in an energy-dependent reaction, a ubiquitin-activating enzyme (E1) activates the ubiquitin and transfers it to a ubiquitin-conjugating enzyme (E2). E2 in turn acts in concert with a third specificity-determining component, E3, to transfer ubiquitin to the target protein. E3s generally function as adapters that link E2s to specific substrates (16, 17). The SCF family of E3 ubiquitin ligases is among the largest and most versatile of the E3s. SCFs are modular complexes that consist of Skp1, a cullin such as Cul1, Rbx1, and one of a large family of substrate adapter proteins, such as F-box proteins. F-box proteins have a second domain that is implicated in substrate recruitment. In SCF complexes, Cul1 (Cdc53 in yeast) serves as a scaffold to bind both to Rbx1 and Skp1. Rbx1 recruits E2, while Skp1 binds the F-box domain of the F-box protein (2, 11, 19, 26, 34, 35, 38, 42). Other SCF-related complexes contain Cul2 and have an Skp1-related component that binds to an F-box-related family of proteins, the BC-box proteins (7, 15). A newly discovered SCF-related complex contains Cul3 and binds to a class of Skp1-F-box fusion proteins, known as BTB proteins, which recruit substrates (28, 41).
It is unclear how cyclin F functions during the cell cycle. Through its cyclin box, it might recruit and activate a Cdk and control proteins through phosphorylation. It might also bind to Cdk inhibitors or other substrates via the cyclin box. Alternatively, through its association with the SCF, cyclin F might direct ubiquitination and destruction of proteins to which it binds or possibly even destruction of itself (43). None of these functions are mutually exclusive.
The ubiquitous nature of cyclin F expression in developing embryos, its tightly cell cycle-regulated expression and phosphorylation, and its high degree of phylogenetic conservation all suggested that regardless of its mode of action, cyclin F is likely to coordinate the timing of critical cell cycle events. In order to investigate these issues, we generated mice that lacked a fully functional cyclin F or that were conditionally deficient for cyclin F. With a number of developmental anomalies due in part to gross defects in placental development, CycF−/− mice die around embryonic day 10.5 (E10.5). Cyclin F is not required for mouse embryonic fibroblast (MEF) viability, although CycF−/− MEFs exhibit a number of defects related to cell cycle progression. Thus, cyclin F is essential for embryonic development and plays a role in cell cycle events.
MATERIALS AND METHOD
Construction of the cyclin F targeting vector and generation of CycF+/− and CycFflox/+ embryonic stem (ES) cells
A 0.6-kb cDNA fragment was used to screen a mouse 129/Sv genomic library. Mapping by standard techniques confirmed this genomic organization. Sequencing exon-intron boundaries based on the predictions from human data confirmed the organization and location of the first six exons of mouse cyclin F. The region indicated in Fig. 1B was digested with SfiI, blunt ended, and digested with XhoI and cloned into pBST via EcoRV-XhoI digestion. A loxP site which contained an EcoRI site was cloned into the SalI site 5′ of exon 2 without conserving the SalI site. A loxP-pGKNEO-loxP cassette was digested with EcoRV-BamHI, end filled, and cloned into the above vector at the PmlI site to complete the targeting construct.
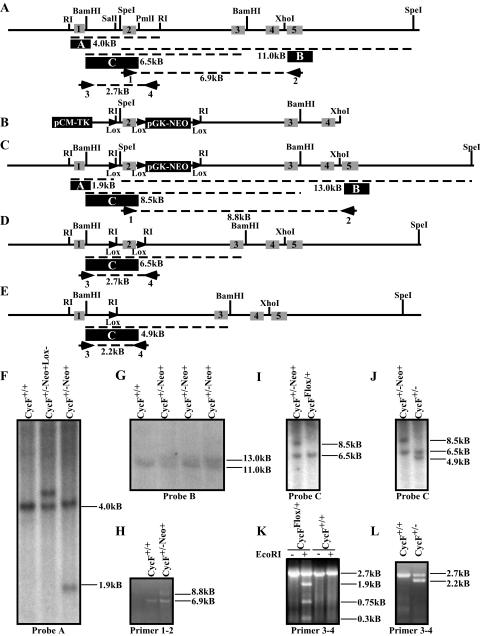
FIG. 1.
Targeted disruption of the mouse cyclin F locus. (A) Restriction map of the cyclin F genomic locus, showing intron-exon boundaries and location for exons 1 to 5 (indicated by grey boxes with black numbers) of mouse cyclin F. The probes used for Southern hybridization are indicated by a filled black box with white letters. Primer pairs used for PCR analysis and mapping are indicated numerically beneath the primer. (B) Map of targeting construct. LoxP sites are indicated by filled triangles. (C) The restriction map of the cyclin F locus following homologous integration of the construct shown in panel B. Predicted PCR fragment sizes are shown. (D) The restriction map of the cyclin F flox allele following Cre-mediated recombination and deletion of the pGK-NEO reporter cassette. (E) The restriction map of the cyclin F deletion allele following Cre-mediated recombination and deletion of the exon 2 and the pGK-NEO reporter cassette. (F) Southern hybridization using probe A (indicated in panel A as a filled black box) following EcoRI digestion of ES cell DNA. (G) Southern hybridization maps the 3′ end of the insertion site using probe B following SpeI digestion of ES cell DNA. (H) PCR analysis of the 3′ end of the targeted cyclin F locus using primer pair 1-2 confirms single homologous integration of the loxP-pGK-NEO-loxP cassette and an otherwise intact 3′ region of the cyclin F locus. (I and J) Southern hybridization using probe C following BamHI digestion of ES cell DNA identifies CycFflox/+ allele (panel I) or CycF+/− (panel J). (K and L) PCR analysis using primer pair 3-4 on ES cell DNA confirms the flox allele by unique sensitivity to EcoRI digestion (panel K) and the deletion allele by its smaller size (panel L). The “−Neo+” designation refers to the cyclin F mutant allele shown in panel C prior to Cre-mediated recombination. The “−Neo+Lox−” designation refers to the allele generated by recombination between the distal loxP site and the loxP-pGKNEO-loxP cassette, so as to exclude the distal loxP site.
AB2.2 129/Sv ES cells were passaged and manipulated on gamma-irradiated feeder cells (SNL 7616) according to standard methods (22). The targeting construct was linearized by restriction digestion by NotI and electroporated into ES cells. These cells were selected for 10 days in G418 (Invitrogen) and ganciclovir (kindly prescribed for research purposes by Eric Tiblier). One hundred ninety-two G418- and ganciclovir-resistant ES cell clones were screened by Southern blot analysis following an EcoRI digest of their genomic DNA using probe A. Seven of these were found to contain the homologously recombined targeting construct. Two of these were further electroporated with pCMV-Cre and plated out at low density (500 to 1,000 cells) on 10-cm feeder dishes. One hundred ninety-two clones were picked from each electroporation and screened by Southern hybridization following BamHI digestion using probe C. One line of the 384 screened was identified by Southern blot analysis and PCR to be CycFflox/+, and 14 were shown to be CycF+/−. One CycFflox/+ and two CycF+/− were injected into C57BL/6 blastocysts, giving rise to 12 chimeric males. These were backcrossed to C57BL/6 females to generate F1 cyclin F+/− animals, and at least one chimera generated from each cell line transmitted the desired allele to its offspring. All genotypes were verified by Expand long-template PCR (Roche) from tail DNA prepared according to standard protocols (22). PCR primers were as follows: CycF#1, 5′-GTGATCCATTGTAGGTGTGCC-3′; CycF#2, 5′-CCTCCACCTTGACCTTAAACGGC-3′; CycF#3, 5′-GGCAGGAGAGGCTAGTGATCTGG-3′; and CycF#4, 5′-GGCTGTTCATCTAGAAGACCAGGG-3′.
Histology, immunohistochemistry, and in situ hybridization
In the case of timed-mating embryos, mice were mated overnight and females were inspected for vaginal plugs the following morning. Noon on the day of plugging was defined as 0.5 day postcoitus. Embryos from timed matings were isolated by standard dissection protocols in Hanks buffered salts solution and Dulbecco's phosphate-buffered saline (Invitrogen). Embryos were fixed in 10% buffered formalin overnight at 4°C. Fixation was followed by dehydration in an ethanol series prior to embedding in paraffin and sectioning. Histological staining was carried out according to standard techniques. For 5-bromo-2′-deoxyuridine (BrdU) labeling, pregnant mothers were injected intraperitoneally at E9.5 with 100 μg of BrdU per g of body weight. Embryos were removed after 1.5 h, fixed, and dehydrated as described above. Small pieces of yolk sacs were reserved for genotyping by PCR. In situ hybridization was performed as described previously (25).
Western blotting
Cell lysates were prepared and immunoblotted as described previously (38). Antibodies used to detect blotted proteins were as follows: anti-cyclin E, anti-cyclin D, anti-cyclin A, and anti-RNR2 (all from Santa Cruz Biotechnologies). Mouse cyclin F antibodies were generated by injecting rabbits with a peptide corresponding to the C terminus of mouse cyclin F and were then affinity purified (Bethyl Laboratories).
MEF derivation, 3T3 protocol, cell culture, serum starvation, and colony formation
MEFs were derived from E13.5 embryos as follows. Crosses between CycFflox/+ and CycF+/− mice generated embryos of all possible cyclin F genotypes. Embryonic heads were reserved for genotyping. Torsos were subjected to trypsinization, mechanical separation, and plating into normal medium (described below). MEFs were immortalized following serial passage of 1.2 × 106 cells per 10-cm dish every 3 days until proliferation was evident (37). Following immortalization, all MEF lines were purified from a single clone. Cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 12% fetal bovine serum (Invitrogen), l-glutamine, nonessential amino acids, and penicillin-streptomycin. Population-doubling times were calculated by plating 2 × 105 immortalized cells per 10-cm dish and counting cell numbers after 72 h. Colony formation assays were performed by plating 7 × 105 MEFs in normal media containing either adenovirus-enhanced green fluorescent protein (Ad-eGFP) or Ad-Cre for 48 h. Following this incubation, MEFs were trypsinized and counted. Five hundred cells per 10-cm dish were plated. After 10 days in culture, colonies were fixed and stained with 2% methylene blue in 70% ethanol solution and counted. Serum starvation experiments were performed as follows. MEFs were grown to confluence in 15-cm dishes and remained in confluence for 72 h. Next, cells were trypsinized, washed in low-serum medium, and plated into 0.1% fetal bovine serum-Du lbecco's modified Eagle's medium at a density of 7 × 105 cells per 10-cm dish for 24 h. Cells were released into normal media and collected at the indicated time points. For fluorescence-activated cell sorter (FACS) analysis, cells were fixed in 70% ethanol solution in phosphate-buffered saline overnight at 4°C and stained with propidium iodide in the presence of RNase for 2 h at 37°C.
RESULT
Generation of mice deficient for cyclin F
To generate the construct for mutating the genomic cyclin F locus in ES cells, we used a 0.6-kb cDNA fragment containing the first five exons of mouse cyclin F to screen a mouse genomic library. A 20-kb fragment containing the 5′ end of the mouse cyclin F locus was isolated. Restriction analysis and sequence data confirmed the genomic organization (Fig. 1A). Because we suspected that cyclin F might be an essential cell cycle gene, we generated a conditional allele, placing a loxP site 300 bp 5′ to exon 2 and a loxP-pGKNEO-loxP cassette 200 bp 3′ to exon 2 (Fig. 1B). The deletion of exon 2 would delete the F box (1, 2) and generate a frame shift that produces a frameshift mutation after a sequence of only 5 amino acids and a premature stop codon after only 39 amino acids. Two splice forms that differ only in the identity of exon 1 have been identified in the database (data not shown). Since all of the known splice forms of cyclin F include exons 2 and 3, this modification is expected to generate a null mutation, barring the existence of as yet unknown splice forms lacking these exons. However, since alternative translational start sites that bypass exon 2 are theoretically possible, we cannot be certain that this allele represents a true null mutation. Our mutant allele, however, is minimally expected to compromise the F box of cyclin F, since this motif is almost entirely encoded by the region deleted.
Mouse cyclin F was disrupted by homologous recombination following electroporation of the targeting vector into murine 129/Sv ES cells. We screened 192 G418- and ganciclovir-resistant clones by Southern hybridization and obtained seven (3.6%) with the appropriate homologously targeted event (Fig. 1A, C, and F). Southern hybridization and PCR verified that a single integration event occurred without otherwise affecting the chromosome (Fig. 1G and H). Cre recombination in ES cells produced a single CycFflox/+ and several CycF+/− ES cell lines, which we verified by Southern hybridization and PCR analysis (Fig. 1I through L). Two separately derived CycF+/− ES cell lines and the single CycFflox/+ ES cell line were injected into donor blastocysts. Chimeric males were backcrossed to C57BL/6 females, and agouti offspring bearing the appropriately targeted chromosome were recovered for each of the three lines injected.
Cyclin F deficiency results in midgestation embryonic lethality
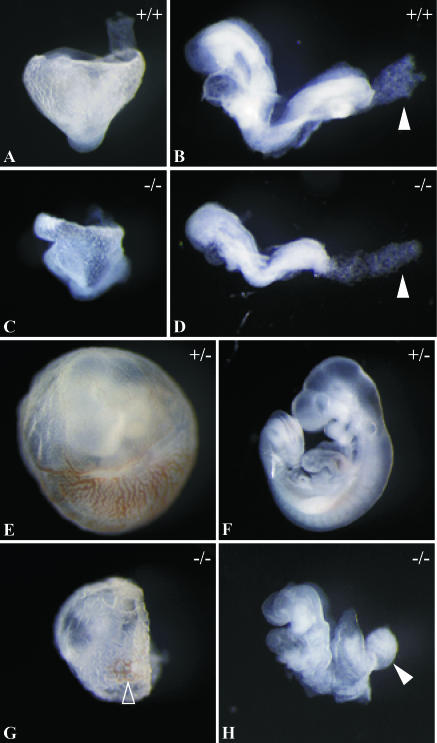
CycF+/− animals appear normal, are fertile, and exhibit no overt phenotypes or spontaneous tumor formation up to 24 months of age. F2 CycF+/− mice were intercrossed to generate CycF−/− animals. However, no CycF−/− animals were recovered from 257 offspring genotyped at the time of weaning (Table 1), suggesting that cyclin F was essential for the completion of embryogenesis. To characterize the timing and quality of the embryonic lethality, we inspected the gross morphology of embryos from timed CycF+/− intercrosses at different stages of development. Embryos were genotyped by yolk sac PCR (Table 1). At E8.5, CycF−/− embryos appeared grossly normal, although they were about 10% smaller than their wild-type littermates (Fig. 2B and D). In all cases, we observed that E8.5 CycF−/− embryos failed to initiate chorio-allantoic fusion. This is evident by the rounded shape of their allantois (Fig. 2D). The mouse allantois gives rise to the future umbilical component of the placenta. Its principal function is to facilitate a connection between the embryonic and maternal circulatory systems for the exchange of nutrients and metabolic wastes. In normal E8.5 embryos, this structure has already fused to the chorion, and as such, manual dissection results in its torn appearance (Fig. 2B). By E9.5, CycF−/− embryos consistently exhibited an array of gross morphological abnormalities (Fig. 2F versus H). CycF−/− embryos are approximately threefold smaller than their wild-type littermates. In 90% of the embryos we analyzed (19 of 21), there was no axial rotation observed whatsoever, whereas normal embryos had turned by this time. CycF−/− embryo bodies appeared kinked, as if axial rotation had begun but failed, thereby twisting the body at the axis about which this rotation occurs. The neural tube of CycF−/− embryos failed to close by E9.5 in all embryos we analyzed, and the leading edges which fuse during closure appeared thickened and kinked. Neural development in CycF−/− embryos was severely retarded as well. Forebrain, midbrain, and hindbrain regions all appeared underdeveloped for gestational age (Fig. 2F versus H). The absence of cyclin F further compromised the development of posterior structures, including the posterior limb bud and the allantois. In CycF−/− embryos, the allantois failed to fuse with the chorion and by E9.5 appears as a bulbous knot (Fig. 2H). Still, many structures normally seen by E9.5, including the first branchial arch, otic cup, eye primordia, and anterior limb bud, appeared to have developed in the CycF−/− embryos. In all cases, CycF−/− embryos possessed a beating heart at E9.5. However, because we never observed live embryos at E10.5 or beyond (Table 1 and data not shown) and because in some cases we observed embryos that had already resorbed at E9.5, we believe that cyclin F-nullizygous embryos die between E9.5 and 10.5.
TABLE 1.
Genotypes of embryos from CycF+/− intercrosses at indicated times
| Age | No. of embryos with CycF genotype:
|
||
|---|---|---|---|
| +/+ | +/− | −/− | |
| Weaning | 82 | 175 | 0 |
| E8.5 | 9 | 13 | 9 |
| E9.5 | 28 | 45 | 21 |
| E10.5 | 4 | 10 | 1a |
Embryo almost completely resorbed.
FIG. 2.
Gross morphology of cyclin F−/− embryos and yolk sacs. Embryos from timed cyclin F+/− intercrosses dissected away from maternal decidua and yolk sacs. Genotyping was confirmed by PCR genotyping of yolk sacs using primer pair 3-4 as shown in Fig. 1. (A) Lateral view of wild-type E8.5 embryo inside the yolk sac. (B) Left-lateral view of wild-type E8.5 embryo. The filled white arrowhead identifies allantois manually torn during dissection. (C) Lateral view of CycF−/− E8.5 embryo inside the yolk sac. (D) Left-lateral view of CycF−/− E8.5 embryo. The filled white arrowhead identifies rounded, unattached allantois. (E) Wild-type embryo inside the yolk sac at E9.5. (F) Left-lateral view of E9.5 CycF+/+ embryo. (G) CycF−/− embryo inside the yolk sac at E9.5. The open arrowhead highlights the area where red hematopoietic tissue is evident. (H) CycF−/− E9.5 embryo. The solid arrowhead indicates bulbous allantois.
Extraembryonic defects in CycF−/− embryos
Mouse models suggest that the disturbances that cause midgestation embryonic lethality often compromise some aspect of the maternal-fetal circulatory exchange (5). CycF−/− embryos are no exception. In addition to the failure of chorioallantoic placental development, the yolk sacs of CycF−/− embryos also appeared abnormal. The mouse yolk sac placenta sustains the majority of maternal-fetal exchange prior to the development of the chorioallantoic placenta and is also the primary site of hematopoiesis. By E8.5, CycF−/− yolk sacs already appeared thin and were more fragile than those of their wild-type littermates (Fig. 2A versus C), and by E9.5, a number of abnormalities emerged. In normal yolk sac development, the blood vessels have a stereotypical branching organization, in which increasingly smaller-diameter, thin-walled vessels are created in order to sustain nutrient exchange, and these vessels are filled with blood cells (Fig. 2E). In CycF−/− E9.5 embryos, the vessels were a disorganized meshwork lacking organized branching patterns, and the vessel walls appeared thicker throughout. Only small amounts of blood were evident in these vessels (Fig. 2G).
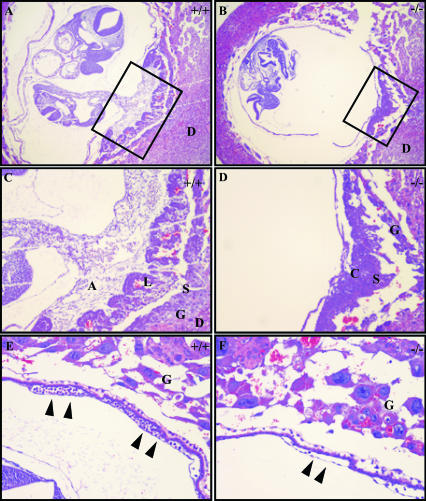
We confirmed the above observations histologically (Fig. 3A and B). Sagittal sections illustrate the architecture of the normal E9.5 placenta. During placental formation, the allantoic vessels invade the chorionic plate, and a loose layer of vascular tissue surrounded by chorionic trophoblasts making up the labyrinthine layer evolves. The extraembryonic spongiotrophoblast and giant cells lie externally to these layers (Fig. 3A and C and 4B). In the most extreme cases in which allantoic fusion and invasion fail completely, the chorion- and spongiotrophoblast cell layers remain condensed, and no labyrinthine maturation is evident (Fig. 3B and D). In other cases, there are changes consistent with labyrinthine maturation, although no physical connection with the allantois is evident in any serial sections taken through these placentas (Fig. 4C and data not shown). Furthermore, histological inspection of the yolk sac revealed defects consistent with those observed grossly. Not only was there a reduction in the number of vessels present in these yolk sacs, but these vessels also contained a distinct paucity of nucleated hematopoietic precursor cells (Fig. 3E and F).
FIG. 3.
Histological examination of CycF−/− placentas and yolk sacs confirms failure of placentation. (A) Shown is ×4 magnification of the wild-type embryo in its yolk sac with functional placental connection. The box indicates the region highlighted in panel C. (B) Shown is ×4 magnification of the CycF−/− embryo in its yolk sac with defective placenta. The box indicates the region highlighted in panel D. (C) Shown is ×10 magnification of the wild-type placenta that illustrates the wild-type E9.5 placenta. (D) The CycF−/− placenta lacks normal architecture and obvious allantois connection. (E) Histological analysis of the normal yolk sac demonstrates abundant large and small vessels with rich blood islands (solid arrowheads). (F) The CycF−/− yolk sac lacks an abundant mature vasculature and lacks a rich nucleated hematopoietic component (solid arrowheads). Abbreviations: A, allantois; L, labyrinthine layer (L); S, spongiotrophoblast layer; G, giant cell layer; D, decidual layer.
FIG. 4.
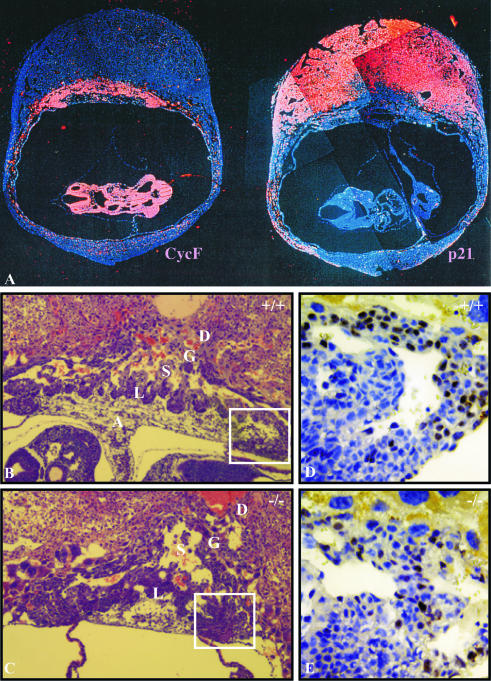
Proliferative defects in CycF−/− placentas. (A) mRNA in situ hybridization on wild-type E9.5 embryos and placentas showing expression of cyclin F (left) and p21(right). Signal appears as red, while nuclei are blue. (B and C) Wild-type and CycF−/− placentas at ×10 magnification. (D and E) Magnification of ×40 illustrates that BrdU-positive cells (brown) of the chorionic trophoblast layer make up roughly 25% of the cells in panel D, compared to around 13% of those in panel E. Sections in panels D and E were counterstained with hematoxylin to produce otherwise blue nuclei. Abbreviations: A, allantois; L, labyrinthine layer; S, spongiotrophoblast layer; G, giant cell layer; D, decidual layer.
Cyclin F expression in extraembryonic tissues correlates with defects in proliferation
We analyzed cyclin F mRNA expression patterns by in situ hybridization in E9.5 embryos. In a pattern similar to that of cyclins A2 and B1, cyclin F was expressed in all actively dividing cells of the mouse embryo (Fig. 4A [left panel] and data not shown). This pattern was mutually exclusive from that of p21 (Fig. 4A [right panel] and data not shown), whose expression generally correlates with cell cycle exit (25). In addition, cyclin F was highly expressed in the chorionic trophoblast layer, where allantoic fusion occurs and labyrinthine maturation commences (Fig. 4A). To assess whether the absence of functional cyclin F correlated with altered cell cycle dynamics in this cell population, we performed BrdU-labeling experiments. There was a distinct reduction in the BrdU incorporation (25 versus 12%) by the trophoblast cells at the chorionic plate adjacent to the area of labyrinthine branching (Fig. 4D versus E). These data suggested that without the function of cyclin F, placental development and maturation failed, and this result was accompanied by the compromised proliferation of distinct populations of trophoblast cells in which cyclin F is normally expressed. Whether the failure in trophoblast proliferation was causal for or merely correlative with the placental failure is not clear.
Tissue-specific insensitivity to cyclin F elimination
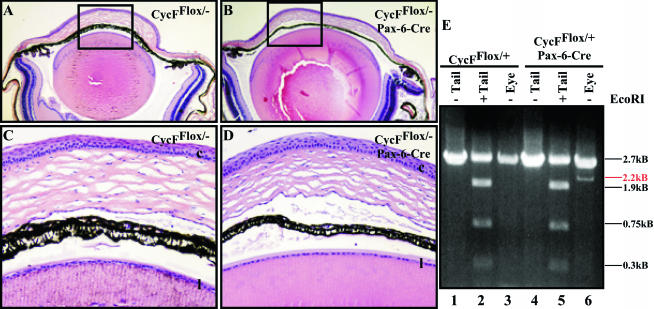
To test whether we were able to identify embryonic or adult tissue with an exquisite sensitivity to cyclin F elimination, we crossed a number of different Cre recombinase driver lines into our CycFflox/− background. Because cyclin F was expressed in all dividing tissues of the embryo and adult, we chose mice expressing Cre in the dividing cells of tissues derived from each of the three embryonic germ layers. The Pax6 promoter we utilized drives the expression of Cre in the ectodermally derived lens and cornea (Paul Overbeek, personal communication). No obvious morphological or histological differences were evident in any of the CycFflox/− mice expressing the Pax-6 Cre transgene (Fig. 5A through D and data not shown). We do not believe that this result was due to the absence of recombination here, since by comparing the genotype of CycFflox/+ tails to eyes in mice with and without the Cre transgene, we were able to demonstrate that Cre did mediate recombination in this tissue (Fig. 5E, lanes 1, 2, and 3 versus 4, 5, and 6). These mice were each of the flox/+ genotype as indicated by their tail DNA; however, the deleted allele emerges in eyes from animals in which the Cre transgene is expressed (Fig. 5E, lane 3 versus lane 6; note the band indicated by the red line). Thus, to the extent that cyclin F is deleted in this tissue, cyclin F appears to be dispensable for cornea and lens development and physiology. Alpha-1-collagen drove Cre expression in the mesodermally derived osteoblast progenitor and osteoblast cells (6). Finally, fatty acid binding protein drove expression of Cre in the endodermally derived distal small intestine, cecum, colon, and bladder (30). None of these tissues showed defects due to the absence of functional cyclin F, indicating that cyclin F is not essential for development and physiology in the bone, gut, and bladder (data not shown).
FIG. 5.
Cyclin F is dispensable for cornea and lens development. (A) Shown is ×4 magnification of flox/− eye section. (B) Shown is ×4 magnification of flox/−;Pax-6-Cre eye section. (C) Shown is ×20x magnification of the boxed region in panel A. (D) Shown is ×20 magnification of the boxed region in panel B. (C and D) Black letters (l and c) represent lens and corneal epithelial layers, respectively. (E) PCR analysis using primer pair 3-4 shows the tail genotype of flox/+ mice. Recombination occurs specifically in the eye of an animal bearing the Pax6-Cre transgene (indicated by the emergence of 2.2-kb band [red line; compare lane 3 to lane 6]).
Cyclin F disruption alters the cell cycle but is not essential for MEF viability
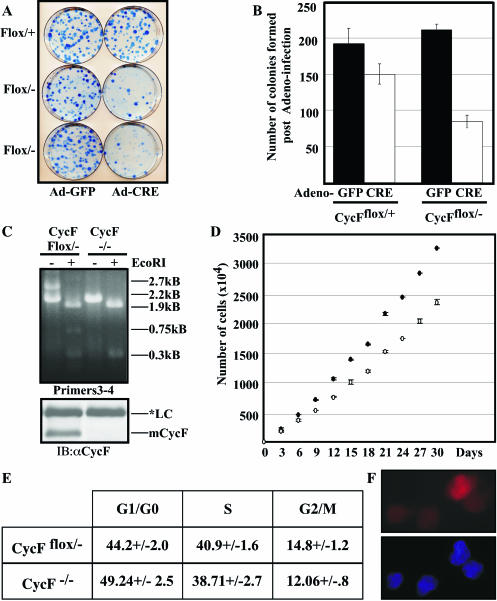
To study the consequence of cyclin F loss from a cellular and biochemical perspective, we derived MEFs that were conditionally deficient for cyclin F (CycFflox/−). Both a CycFflox/− and a CycFflox/+ control were subjected to immortalization through the 3T3 protocol (37). We then subjected these lines to Cre-mediated recombination via Ad-Cre infection, using Ad-GFP as a control, and we measured the respective capacities to form colonies following low-density plating. We observed a marked reduction in both the quality and quantity of macroscopic colony formation (Fig. 6A and B). While Ad-Cre infection in control cells did result in a small reduction in the number of colonies, both the quantitative and qualitative differences in the CycFflox/− lines following Ad-Cre infection were far more dramatic than those observed in the CycFflox/+ controls. For the purposes of quantification (Fig. 6B), we counted all colonies that were macroscopically visible and did not make any distinction based on colony size per se. These data are consistent with a defect in cell proliferation and not with incomplete infection for the following reasons. First, no residual floxed allele and accompanying full-length cyclin F protein were evident by PCR and Western blot analysis, respectively, in the bulk population of MEFs following Ad-Cre infection prior to colony plating (data not shown). Second, colonies that did grow from the Ad-Cre-infected plates were isolated and characterized, and we ascertained that in every case, these represented CycF−/− outgrowths (Fig. 6C). Furthermore, these CycF−/− cell lines also exhibited defective colony formation in subsequent experiments (data not shown).
FIG. 6.
Cell cycle defects in MEFs lacking cyclin F. (A) Colony formation defects in CycFflox/− MEFs (bottom two rows of plates) following Ad-Cre versus Ad-GFP infection compared to similar infections of CycFflox/+ controls. (B) Quantification of colonies shown in panel A. (C) PCR (top) and Western blot (bottom) analyses confirm the deletion of exon 2 and the absence of full-length cyclin F protein, respectively, in clones derived from CycFflox/− MEFs following Ad-Cre infection. Immunoblotting (IB) was performed with an anti-CycF antibody (αCycF). LC, loading control. (D) Population-doubling defects in CycF−/− MEFs (open circles) compared to those of their CycFflox/− parents (filled circles). (E) Summary of asynchronous FACS profiles for CycFflox/− parent and CycF−/− derivatives. (F) Cyclin B (red) localizes to the nucleus in the absence of cyclin F (top panel); 4′,6′-diamidino-2-phenylindole (DAPI) (blue) stain confirms the location of the nucleus.
The fact that we were able to culture CycF−/− MEFs is consistent with our previous conclusions regarding the tissue insensitivity to cyclin F elimination. Namely, while cyclin F is essential for the completion of embryogenesis, it is dispensable for cellular viability and proliferation in MEFs. Consistent with this conclusion and contrary to previous reports demonstrating a supposed dependence on cyclin F for the appropriate nuclear localization of cyclin B1 (20), we observed efficient nuclear access of cyclin B1 despite the absence of cyclin F (Fig. 6F).
We also studied the gross doubling times of asynchronously growing CycF−/− MEFs compared to those of the CycFflox/− parent lines from which they were derived. Over the course of 30 days, the CycFflox/− parent lines grew roughly 1.1 times faster than their CycF−/− derivatives, again consistent with slight defects in MEF proliferation in the absence of cyclin F (Fig. 6D). Analogous results were obtained on shorter time courses for other CycF−/− MEF lines. When we assessed these asynchronous cultures for their respective cell cycle distributions, we observed a subtle, yet statistically significant, difference (Fig. 6E). Namely, there was a 5% increase in the number of cells in G1/G0 and a 2.2% decrease in the number of cells in G2/M (Fig. 6E). These data represent the summary of six independent collections of asynchronous cell cultures of the indicated genotypes.
Cyclin F−/− MEFs display slowed cell cycle reentry
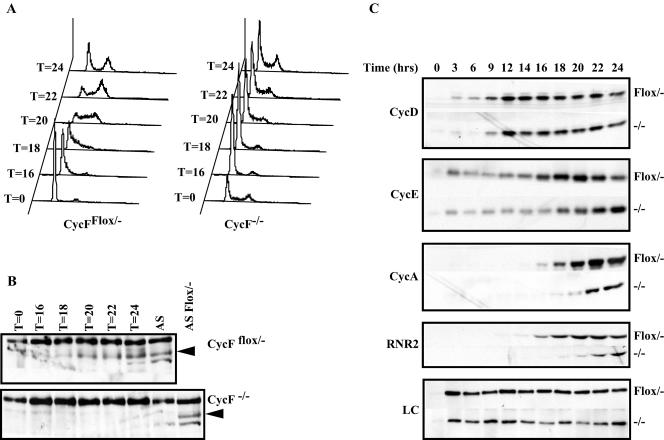
To identify the timing and nature of the cell cycle defect of CycF−/− MEFs, we subjected them and their CycFflox/− parents to serum starvation to arrest them in the G0 quiescent state and stimulated them by serum addition to reenter the cell cycle. We monitored their progression through the cell cycle by FACS analysis and observed that CycF−/− cells were significantly delayed in their capacity to reenter the cell cycle (Fig. 7A). At 20 h following serum addition, almost 80% of the CycF−/− cell population remained in G0/G1, whereas nearly the entire control cell population had entered the cell cycle (Fig. 7A). Similar results were obtained in other experiments comparing CycFflox/− lines following Ad-GFP versus Ad-Cre infection (data not shown). Thus, we do not believe that this phenomenon was simply the consequence of clonal variation. This delay either represented a global failure in the induction of cell cycle-associated molecules or a specific delay in cell cycle progression in the face of normal temporal expression of cell cycle-regulated gene products. To distinguish between these possibilities, we assessed the expression profile of various cell cycle-specific markers, i.e., cyclin D1, cyclin E1, cyclin A1, and the R2 subunit of the ribonucleotide reductase enzyme (RNR2). While cyclin D expression in CycFflox/− MEFs is evident 6 h earlier than in their CycF−/− derivatives, its expression peaks at approximately the same time following serum readdition in both cell lines (Fig. 7C, top panel). However, both the induction and peak expression profiles are substantially more delayed for all the other cell cycle markers, a finding that is consistent with a specific cell cycle defect in CycF−/− MEFs occurring between the induction of cyclin D and cyclin E.
FIG. 7.
Absence of cyclin F slows cell cycle reentry following serum starvation between the induction of cyclins D and E. (A) FACS profiles of CycFflox/− (left) and CycF−/− (right) cells following serum starvation and stimulation following serum addition. Times are indicated in hours. (B) Western blot analysis using cyclin F antibodies confirms the temporal regulation of cyclin F expression during the cell cycle. (C) Shown are analyses similar to those in panel B for cyclin D1, cyclin E1, cyclin A, and RNR2 that illustrate the delay in peak cyclin E, cyclin A, and RNR2 induction compared to that of cyclin D. Loading control (LC) is a nonspecific band recognized by cyclin F antibodies. For each blot, the genotype is indicated to the right, and the employed antibody is indicated to the left.
DISCUSSION
Cyclin F is a phylogenetically conserved cell cycle-regulated protein implicated in cell cycle control and possibly proteolysis through its ability to bind the Skp1 protein, a central component of the SCF ubiquitin ligase complex. To explore cyclin F function, both in cells and during the development of an organism, we generated mice wholly deficient or conditionally deficient for cyclin F expression. Cyclin F is required for the successful completion of embryogenesis. CycF−/− embryos exhibit a wide array of developmental problems, including a failure to complete axial rotation and defects and/or delays in neural tube closure and brain development. Both the yolk sac and chorioallantoic placenta fail to develop normally in the absence of cyclin F (see below). CycF−/− MEFs were derived to enable us to study the cell cycle defects associated with the absence of cyclin F function. Our analyses of these cells revealed that, while cyclin F is essential for the completion of embryogenesis, fully functional cyclin F is dispensable for cell division under serum-rich (i.e., stimulatory) conditions. In addition, cyclin F is important for cell cycle reentry out of quiescence.
Cyclin F is essential for the completion of embryogenesis
CycF−/− embryos die prior to E10.5 due to an array of developmental anomalies and delays in the embryo proper as well as in the extraembryonic tissues. Thus, like cyclin A2 (23) and cyclin B1 (3), cyclin F joins the class of cyclins whose functions cannot normally be compensated for by the other cyclins of the same or related classes (reviewed in reference 39). The closely related A-type cyclins, which are expressed in a completely overlapping pattern with cyclin F both in the embryo and during the cell cycle and which are the most closely related in the cyclin box region to cyclin F, cannot compensate for its function. Our results also distinguish cyclin F from G1 cyclins. Animals carrying single and double mutations of D-type cyclins produce only cell type-specific defects likely due to the inability of discrete cell types to mobilize the expression of the remaining cyclins of that class, but such mutations do not result in early embryonic lethality (4, 10, 32, 33). Similarly, cyclin E single mutants complete embryogenesis normally (13).
Although we have not formally ruled out the possibility that cyclin F is essential for the development of discrete tissue types, it is possible that the embryonic defects observed in CycF−/− embryos are secondary consequences of nutrient compromise due to the complete failure of yolk sac and chorio-allantoic placentation. We observed frank defects in the maturation of the yolk sac and chorioallantoic placenta in precisely the locations where cyclin F is expressed. Other cyclins and cyclin-regulated molecules whose principal embryonic functions appear to be placental development include the E-type cyclins (13) and the retinoblastoma protein (40). In addition, tissue-specific elimination of cyclin F in the lens, cornea, bone, jejunum, ileum, cecum, colon, and bladder revealed no specific cellular sensitivity to cyclin F function during embryogenesis or adulthood. For these reasons, we feel that nutrient deprivation imposed by the placental defects may be the primary cause of the defects observed during embryogenesis. This hypothesis will need be tested in future experiments through tetraploid rescue or the employment of Mox2+/Cre transgenics, which express Cre only in embryonic tissues (18, 36).
Cyclin F is not essential for MEF viability but does affect the MEF cell cycle
Our cellular analyses demonstrated that cyclin F is not essential for MEF viability. While the elimination of cyclin F compromised the ability to form colonies following low-density plating, the size of these colonies was more severely affected than was the overall number of colonies. This finding is consistent with the delays in doubling time observed for CycF−/− MEFs and suggests that the inability to form colonies is due to the inability of CycF−/− MEFs to exit their pseudoquiescent state induced by low-density plating. Cyclin F appears to be dispensable for cyclin B's nuclear translocation, ruling out cyclin F as the sole mediator of this process, as was previously proposed (20).
Previous analyses (1) and results described herein (Fig. 7B) showed that cyclin F expression is tightly cell cycle regulated, beginning in S phase, peaking in late G2, and declining sharply as cells enter mitosis. However, CycF−/− MEFs exhibited cell cycle abnormalities and delays in cell cycle progression that were consistent with cyclin F playing a role in cell cycle reentry and the traversal of the G0/G1 boundary, possibly G1/S. Our results are consistent with the model that the absence of a fully functional cyclin F compromises this process in a specific way rather than generally altering the normal temporal pattern of gene expression for the following reasons. While the temporal expression of most of the cell cycle-regulated markers we analyzed was altered, cyclin D expression, while slightly temporally delayed, peaked identically in both CycF−/− and CycFflox/− parent lines. This result suggests that the inability to reenter the cell cycle in the absence of CycF occurs between the induction of cyclin D and cyclin E. Consistent with these in vitro observations, the absence of cyclin F in the chorionic trophoblast cells of the placenta produces cell cycle defects similar to those observed in MEFs lacking cyclin F. Namely, there is an apparent reduction in the number of these cells entering S phase. Thus, we feel that a likely role for cyclin F is to execute a function in S/G2 that has a positive impact on subsequent G0/G1 or G1/S transitions. Since cyclin D induction appears relatively unperturbed (but a significant delay is observed in cyclin E accumulation), the most likely part of the cell cycle affected is the pathway leading to cyclin E expression. This involves the retinoblastoma (Rb) family of pocket proteins, the E2F proteins, and possibly the kinase activity of cyclin D/CDK4/6 itself. We did not observe an increase in the abundance of p21 or p27 in these cells (unpublished data).
The results of our studies are consistent with cyclin F playing a role, in part, to generate a positively acting signal that persists into the next cell cycle to promote S-phase entry. Alternatively, and perhaps more likely, cyclin F might act to destroy an inhibitory factor during S/G2 whose presence in the next cycle interferes in part with S-phase entry, possibly through cyclin F's capacity as an SCF component. These possibilities cannot be distinguished at the present time. Cyclin F may also normally play a role in G1. While its expression timing in MEFs and HeLa cells suggest an S/G2 expression pattern, it does parallel cyclin A expression, and cyclin A has been implicated in G1/S transitions in other cell types (14). It is also possible that the temporal pattern of cyclin F expression differs in vivo relative to that observed in vitro.
The notion that cyclin F promotes function of the Rb-E2F-cyclin E axis is consistent with the observed defects in trophoblast proliferation. Trophoblasts are the most sensitive tissue to Rb function. Loss of Rb causes a specific hyperproliferation of those cells during development, leading to placental failure (40). It would not be surprising that these cells would also be very sensitive to increased Rb function or decreased E2F-cyclin E function. The latter would result in the reduced proliferation of chorionic trophoblast cells that normally undergo multiple rounds of division during placentation.
Cyclin F joins cyclins A2 and B1 as the only cyclins that are essential for embryonic development. Unlike these cyclins, the loss of which causes cell lethality, cyclin F is required only for development. While cyclin F had been previously implicated in control of cyclin B localization, this was not found to be the case through our genetic studies. It is more likely that among cyclin F's functions is a role in maintaining the homeostasis of other cell cycle regulators that are important for the ability of cells to traverse the G0/G1 or G1/S boundary. The generation of the cyclin F-deficient and cyclin F conditionally deficient animals and cells should help further elucidate these functions in the future.
Acknowledgments
We thank D. Killen for help with sectioning and A. Major for anti-BrdU stains. M.T.T. thanks A. Osborn, J. T. Winston, D. Cortez, D. Schmucker, D. Killen, and T. Westbrook for helpful discussions throughout the course of this work. We are especially grateful to Paul Overbeek for generously providing Pax6-Cre mice prior to publication and for helpful analysis of conditional knockout mice.
M.T.T. was supported by NIH/NIDDK training grant number T32DK07696. This work was supported by NIH grant numbers AG11085 and CA58024 (to J.W.H. and S.J.E.) and DK57693 (to K.M.). S.J.E. is an Investigator with the Howard Hughes Medical Institute.
REFERENCE
- 1.Bai, C., R. Richman, and S. J. Elledge. 1994. Human cyclin F. EMBO J. 13:6087-6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bai, C., P. Sen, K. Hofmann, L. Ma, M. Goebl, J. W. Harper, and S. J. Elledge. 1996. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86:263-274. [DOI] [PubMed] [Google Scholar]
- 2a.Bai, C. 1996. Ph.D. thesis. Baylor College of Medicine, Houston, Tex.
- 3.Brandeis, M., I. Rosewell, M. Carrington, T. Crompton, M. A. Jacobs, J. Kirk, J. Gannon, and T. Hunt. 1998. Cyclin B2-null mice develop normally and are fertile whereas cyclin B1-null mice die in utero. Proc. Natl. Acad. Sci. USA 95:4344-4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ciemerych, M. A., A. M. Kenney, E. Sicinska, I. Kalaszczynska, R. T. Bronson, D. H. Rowitch, H. Gardner, and P. Sicinski. 2002. Development of mice expressing a single D-type cyclin. Genes Dev. 16:3277-3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Copp, A. J. 1995. Death before birth: clues from gene knockouts and mutations. Trends Genet. 11:87-93. [DOI] [PubMed] [Google Scholar]
- 6.Dacquin, R., M. Starbuck, T. Schinke, and G. Karsenty. 2002. Mouse α1(I)-collagen promoter is the best known promoter to drive efficient Cre recombinase expression in osteoblast. Dev. Dyn. 224:245-251. [DOI] [PubMed] [Google Scholar]
- 7.Deshaies, R. J. 1999. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15:435-467. [DOI] [PubMed] [Google Scholar]
- 8.Draviam, V. M., S. Orrechia, M. Lowe, R. Pardi, and J. Pines. 2001. The localization of human cyclins B1 and B2 determines CDK1 substrate specificity and neither enzyme requires MEK to disassemble the Golgi apparatus. J. Cell Biol. 152:945-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evans, T., E. T. Rosenthal, J. Youngblom, D. Distel, and T. Hunt. 1983. Cyclin: a protein specified by maternal mRNA in sea urchin eggs that is destroyed at each cleavage division. Cell 33:389-396. [DOI] [PubMed] [Google Scholar]
- 10.Fantl, V., G. Stamp, A. Andrews, I. Rosewell, and C. Dickson. 1995. Mice lacking cyclin D1 are small and show defects in eye and mammary gland development. Genes Dev. 9:2364-2372. [DOI] [PubMed] [Google Scholar]
- 11.Feldman, R. M., C. C. Correll, K. B. Kaplan, and R. J. Deshaies. 1997. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell 91:221-230. [DOI] [PubMed] [Google Scholar]
- 12.Geng, Y., W. Whoriskey, M. Y. Park, R. T. Bronson, R. H. Medema, T. Li, R. A. Weinberg, and P. Sicinski. 1999. Rescue of cyclin D1 deficiency by knockin cyclin E. Cell 97:767-777. [DOI] [PubMed] [Google Scholar]
- 13.Geng, Y., Q. Yu, E. Sicinska, M. Das, J. E. Schneider, S. Bhattacharya, W. M. Rideout, R. T. Bronson, H. Gardner, and P. Sicinski. 2003. Cyclin E ablation in the mouse. Cell 114:431-443. [DOI] [PubMed] [Google Scholar]
- 14.Guadagno, T. M., M. Ohtsubo, J. M. Roberts, and R. K. Assoian. 1993. A link between cyclin A expression and adhesion-dependent cell cycle progression. Science 262:1572-1575. [DOI] [PubMed] [Google Scholar]
- 15.Harper, J. W., J. L. Burton, and M. J. Solomon. 2002. The anaphase-promoting complex: it's not just for mitosis any more. Genes Dev. 16:2179-2206. [DOI] [PubMed] [Google Scholar]
- 16.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 17.Hershko, A., H. Heller, S. Elias, and A. Ciechanover. 1983. Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J. Biol. Chem. 258:8206-8214. [PubMed] [Google Scholar]
- 18.James, R. M., A. H. Klerkx, M. Keighren, J. H. Flockhart, and J. D. West. 1995. Restricted distribution of tetraploid cells in mouse tetraploid↔diploid chimaeras. Dev. Biol. 167:213-226. [DOI] [PubMed] [Google Scholar]
- 19.Kamura, T., D. M. Koepp, M. N. Conrad, D. Skowyra, R. J. Moreland, O. Iliopoulos, W. S. Lane, W. G. Kaelin, Jr., S. J. Elledge, R. C. Conaway, J. W. Harper, and J. W. Conaway. 1999. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science 284:657-661. [DOI] [PubMed] [Google Scholar]
- 20.Kong, M., E. A. Barnes, V. Ollendorff, and D. J. Donoghue. 2000. Cyclin F regulates the nuclear localization of cyclin B1 through a cyclin-cyclin interaction. EMBO J. 19:1378-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, D., M. M. Matzuk, W. K. Sung, Q. Guo, P. Wang, and D. J. Wolgemuth. 1998. Cyclin A1 is required for meiosis in the male mouse. Nat. Genet. 20:377-380. [DOI] [PubMed] [Google Scholar]
- 22.McMahon, A. P., and A. Bradley. 1990. The Wnt-1 (int-1) proto-oncogene is required for development of a large region of the mouse brain. Cell 62:1073-1085. [DOI] [PubMed] [Google Scholar]
- 23.Murphy, M., M. G. Stinnakre, C. Senamaud-Beaufort, N. J. Winston, C. Sweeney, M. Kubelka, M. Carrington, C. Brechot, and J. Sobczak-Thepot. 1997. Delayed early embryonic lethality following disruption of the murine cyclin A2 gene. Nat. Genet. 15:83-86. [DOI] [PubMed] [Google Scholar]
- 24.Parisi, T., A. R. Beck, N. Rougier, T. McNeil, L. Lucian, Z. Werb, and B. Amati. 2003. Cyclins E1 and E2 are required for endoreplication in placental trophoblast giant cells. EMBO J. 22:4794-4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parker, S. B., G. Eichele, P. Zhang, A. Rawls, A. T. Sands, A. Bradley, E. N. Olson, J. W. Harper, and S. J. Elledge. 1995. p53-independent expression of p21Cip1 in muscle and other terminally differentiating cells. Science 267:1024-1027. [DOI] [PubMed] [Google Scholar]
- 26.Patton, E. E., A. R. Willems, D. Sa, L. Kuras, D. Thomas, K. L. Craig, and M. Tyers. 1998. Cdc53 is a scaffold protein for multiple Cdc34/Skp1/F-box protein complexes that regulate cell division and methionine biosynthesis in yeast. Genes Dev. 12:692-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pines, J. 1996. Cyclin from sea urchins to HeLas: making the human cell cycle. Biochem. Soc. Trans. 24:15-33. [DOI] [PubMed] [Google Scholar]
- 28.Pintard, L., J. H. Willis, A. Willems, J. L. Johnson, M. Srayko, T. Kurz, S. Glaser, P. E. Mains, M. Tyers, B. Bowerman, and M. Peter. 2003. The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase. Nature 425:311-316. [DOI] [PubMed] [Google Scholar]
- 29.Rosenthal, E. T., T. Hunt, and J. V. Ruderman. 1980. Selective translation of mRNA controls the pattern of protein synthesis during early development of the surf clam, Spisula solidissima. Cell 20:487-494. [DOI] [PubMed] [Google Scholar]
- 30.Saam, J. R., and J. I. Gordon. 1999. Inducible gene knockouts in the small intestinal and colonic epithelium. J. Biol. Chem. 274:38071-38082. [DOI] [PubMed] [Google Scholar]
- 31.Schwob, E., T. Bohm, M. D. Mendenhall, and K. Nasmyth. 1994. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell 79:233-244. [DOI] [PubMed] [Google Scholar]
- 32.Sicinski, P., J. L. Donaher, Y. Geng, S. B. Parker, H. Gardner, M. Y. Park, R. L. Robker, J. S. Richards, L. K. McGinnis, J. D. Biggers, J. J. Eppig, R. T. Bronson, S. J. Elledge, and R. A. Weinberg. 1996. Cyclin D2 is an FSH-responsive gene involved in gonadal cell proliferation and oncogenesis. Nature 384:470-474. [DOI] [PubMed] [Google Scholar]
- 33.Sicinski, P., J. L. Donaher, S. B. Parker, T. Li, A. Fazeli, H. Gardner, S. Z. Haslam, R. T. Bronson, S. J. Elledge, and R. A. Weinberg. 1995. Cyclin D1 provides a link between development and oncogenesis in the retina and breast. Cell 82:621-630. [DOI] [PubMed] [Google Scholar]
- 34.Skowyra, D., K. L. Craig, M. Tyers, S. J. Elledge, and J. W. Harper. 1997. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91:209-219. [DOI] [PubMed] [Google Scholar]
- 35.Skowyra, D., D. M. Koepp, T. Kamura, M. N. Conrad, R. C. Conaway, J. W. Conaway, S. J. Elledge, and J. W. Harper. 1999. Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science 284:662-665. [DOI] [PubMed] [Google Scholar]
- 36.Tallquist, M. D., and P. Soriano. 2000. Epiblast-restricted Cre expression in MORE mice: a tool to distinguish embryonic vs. extra-embryonic gene function. Genesis 26:113-115. [DOI] [PubMed] [Google Scholar]
- 37.Todaro, G. J., and H. Green. 1963. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol. 17:299-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winston, J. T., P. Strack, P. Beer-Romero, C. Y. Chu, S. J. Elledge, and J. W. Harper. 1999. The SCFβ-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IκBα and β-catenin and stimulates IκBα ubiquitination in vitro. Genes Dev. 13:270-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Winston, N. 2001. Regulation of early embryo development: functional redundancy between cyclin subtypes. Reprod. Fertil. Dev. 13:59-67. [DOI] [PubMed] [Google Scholar]
- 40.Wu, L., A. de Bruin, H. I. Saavedra, M. Starovic, A. Trimboli, Y. Yang, J. Opavska, P. Wilson, J. C. Thompson, M. C. Ostrowski, T. J. Rosol, L. A. Woollett, M. Weinstein, J. C. Cross, M. L. Robinson, and G. Leone. 2003. Extra-embryonic function of Rb is essential for embryonic development and viability. Nature 421:942-947. [DOI] [PubMed] [Google Scholar]
- 41.Xu, L., Y. Wei, J. Reboul, P. Vaglio, T. H. Shin, M. Vidal, S. J. Elledge, and J. W. Harper. 2003. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature 425:316-321. [DOI] [PubMed] [Google Scholar]
- 42.Zheng, N., B. A. Schulman, L. Song, J. J. Miller, P. D. Jeffrey, P. Wang, C. Chu, D. M. Koepp, S. J. Elledge, M. Pagano, R. C. Conaway, J. W. Conaway, J. W. Harper, and N. P. Pavletich. 2002. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 416:703-709. [DOI] [PubMed] [Google Scholar]
- 43.Zhou, P., and P. M. Howley. 1998. Ubiquitination and degradation of the substrate recognition subunits of SCF ubiquitin-protein ligases. Mol. Cell 2:571-580. [DOI] [PubMed] [Google Scholar]