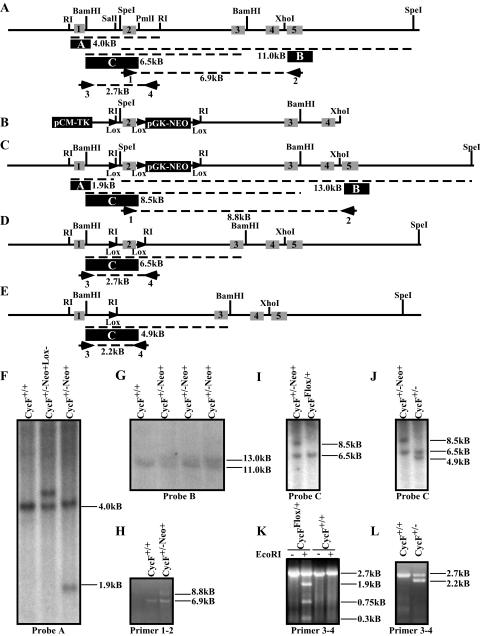
FIG. 1.
Targeted disruption of the mouse cyclin F locus. (A) Restriction map of the cyclin F genomic locus, showing intron-exon boundaries and location for exons 1 to 5 (indicated by grey boxes with black numbers) of mouse cyclin F. The probes used for Southern hybridization are indicated by a filled black box with white letters. Primer pairs used for PCR analysis and mapping are indicated numerically beneath the primer. (B) Map of targeting construct. LoxP sites are indicated by filled triangles. (C) The restriction map of the cyclin F locus following homologous integration of the construct shown in panel B. Predicted PCR fragment sizes are shown. (D) The restriction map of the cyclin F flox allele following Cre-mediated recombination and deletion of the pGK-NEO reporter cassette. (E) The restriction map of the cyclin F deletion allele following Cre-mediated recombination and deletion of the exon 2 and the pGK-NEO reporter cassette. (F) Southern hybridization using probe A (indicated in panel A as a filled black box) following EcoRI digestion of ES cell DNA. (G) Southern hybridization maps the 3′ end of the insertion site using probe B following SpeI digestion of ES cell DNA. (H) PCR analysis of the 3′ end of the targeted cyclin F locus using primer pair 1-2 confirms single homologous integration of the loxP-pGK-NEO-loxP cassette and an otherwise intact 3′ region of the cyclin F locus. (I and J) Southern hybridization using probe C following BamHI digestion of ES cell DNA identifies CycFflox/+ allele (panel I) or CycF+/− (panel J). (K and L) PCR analysis using primer pair 3-4 on ES cell DNA confirms the flox allele by unique sensitivity to EcoRI digestion (panel K) and the deletion allele by its smaller size (panel L). The “−Neo+” designation refers to the cyclin F mutant allele shown in panel C prior to Cre-mediated recombination. The “−Neo+Lox−” designation refers to the allele generated by recombination between the distal loxP site and the loxP-pGKNEO-loxP cassette, so as to exclude the distal loxP site.