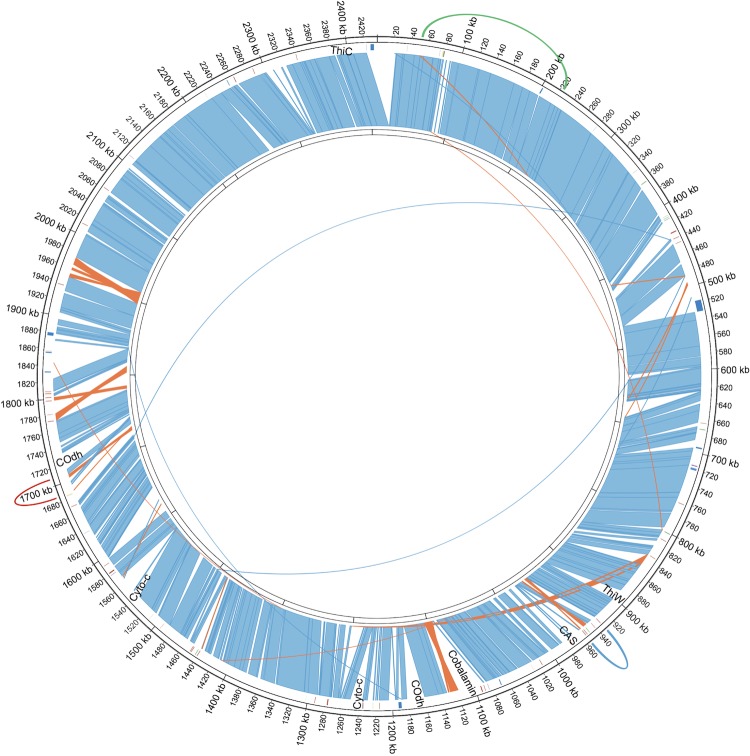
Figure 2.
Genomic alignment of P. oguniense with P. arsenaticum. Outer ring: P. oguniense (+ strand); Inner ring: P. arsenaticum (- strand). Inter-species alignment blocks shown in light blue and gold (inverted orientation). Intra-species P. oguniense genomic inversions shown as arcs of different colors along outer ring: red: C8 inversion (red); Glutamate Dehydrogenase (GluDH) inversion (green); RAMP/paREP inversion (blue). Positions of paREP elements shown as ticks inside outer ring: paREP1 (red); paREP2b (blue); paREP7 (green). Positions of selected genes which are present in P. oguniense and missing in P. arsenaticum are shown in text inside outer ring: thiamine biosynthesis genes (ThiW and ThiC); CRISPR Cassette(CAS); cobalamin cluster; CO dehydrogenase(COdh); and the aerobic cytochrome clusters(Cyto-c). Aligned regions smaller than 500 nucleotides have been removed for clarity.