Figure 3. .
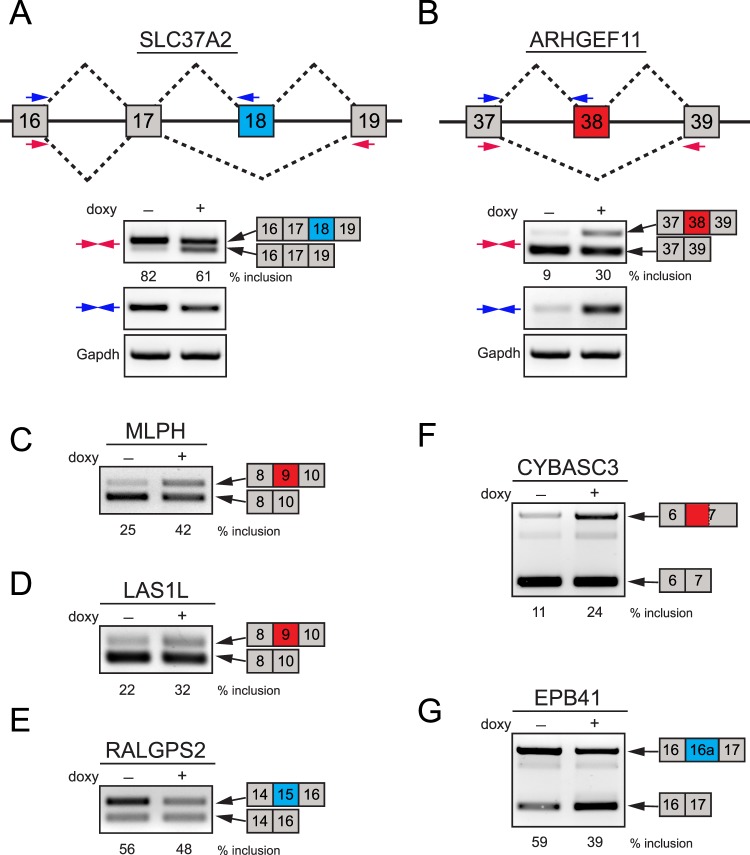
Examples of validated ESRP1-regulated splicing events in HCET cells. (A, B) Two examples of alternatively spliced genes in ESRP1 knockdown HCET cells are presented. Diagrams show the structure of exons that are alternatively spliced in SLC37A2 and ARHGEF11. The location of two primer sets (colored arrows) used for RT-PCR validation for each gene is indicated. While SLC37A2 exhibited decreased inclusion of exon 18 (shown in blue), ARHGEF11 specifically displayed increased inclusion of exon 38 following ESRP1 knockdown. Inclusion percentages of alternative exons are shown for each condition. (C–E) Three examples of known ESRP-target events validated in ESRP1 knockdown HCET cells are shown. Alternative cassette exons of MLPH, LAS1L, and RALGPS2 showed specific changes in inclusion rate upon ESRP1 knockdown. (F) An example of increased alternative 3′ splice site usage is shown. Upon ESRP1 knockdown, CYBASC2 displayed enhanced usage of alternative 3′ splice site (779 bp upstream of major splice site of exon 7). (G) A previously unidentified ESRP1-regulated cassette exon in EPB41 is validated in HCET cells. Exon 16a of EPB41 showed decreased inclusion upon ESRP1 knockdown. All alternatively spliced PCR products shown in Figure 3 were verified through sequencing.