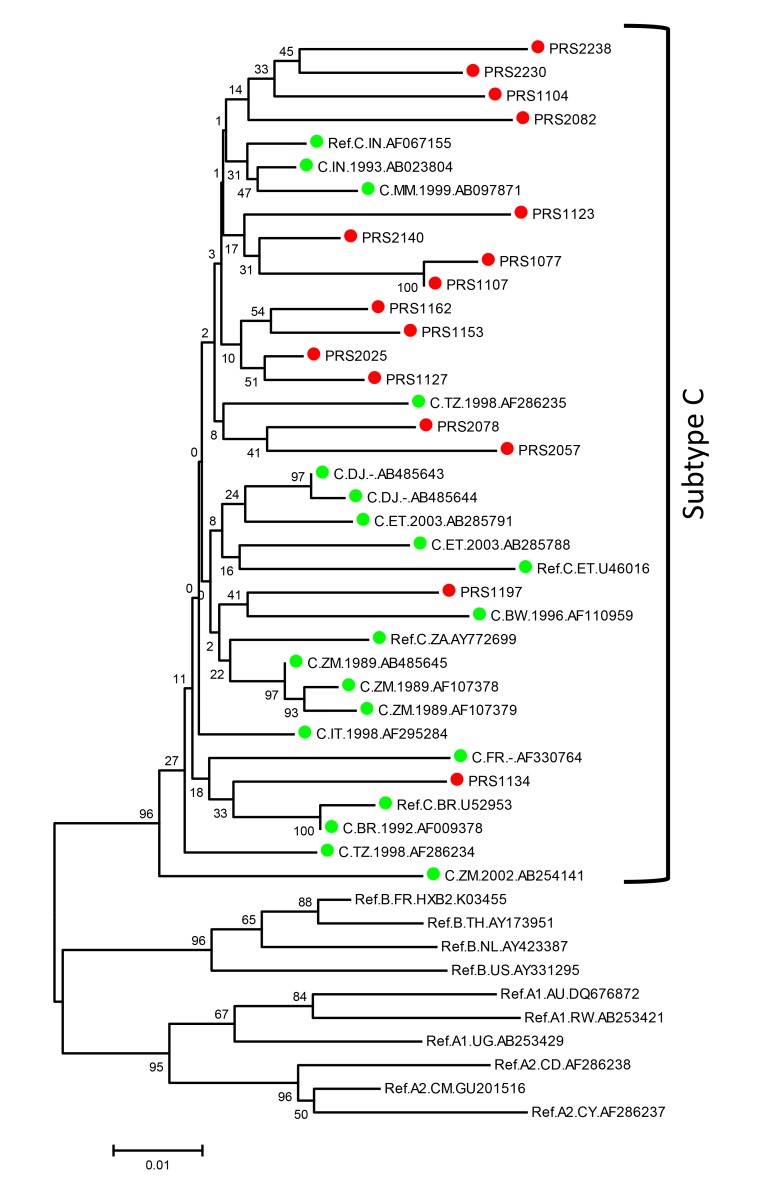
Figure 1. Phylogenetic analysis for characterisation of HIV-1 subtypes: The phylogenetic tree was constructed using general time reversible (GTR) model with gamma distribution and invariant sites (GTR+G+I) as observed best fitted model for the dataset with cohort sequences and reference sequences downloaded from Los Alamos Database (www.hiv.lanl.gov).
Subtype C reference sequences from different countries are marked with green filled circle. Cohort sequences are shown with red filled circle.