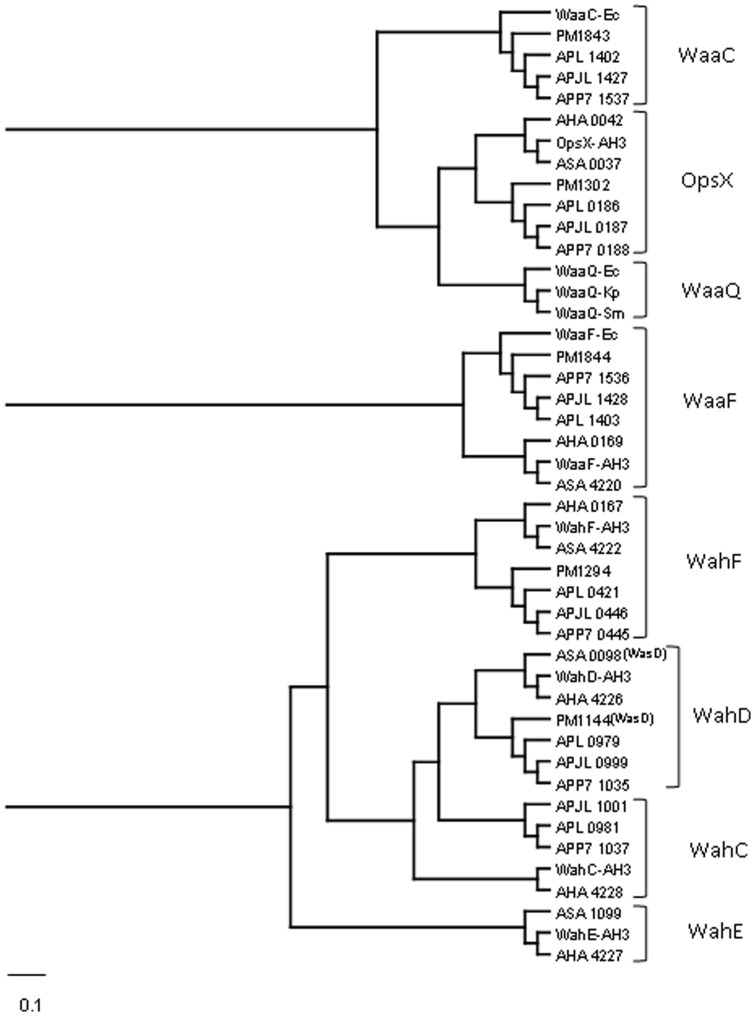
Figure 2. Cladogram obtained running the phylogenetic inference software Protpars from the PHYLIP package version 3.5c as indicated in Materials and Methods section.
The separation and the large of the lanes are relative to the similarity degree according to the program used (the scale bar indicates an evolutionary distance of 0.1 aminoacid substitutions per position).The cladogram shows the similarity relationship among putative heptosyltransferases from A. pleunopneumoniae strains L20, JL03, and AP76 (APL, APJL, and APP7, respectively), A. hydrophila AH3 and ATCC7966T strains (AH-3 and AHA, respectively), A. salmonicida A450 strain (ASA), E. coli (Ec), K. pneumoniae (Kp), S. marcescens (Sm), and P. multocida (PM).