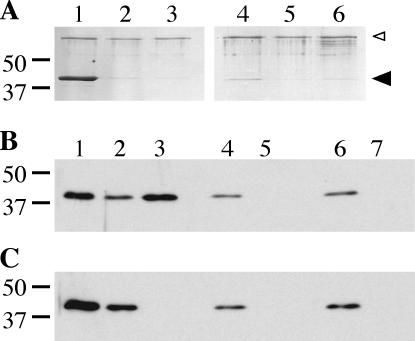
FIG. 8.
Nucleotide binding capacity of DotB and DotB K162Q. (A) Coomassie-stained SDS-PAGE gel showing elution fractions from DotB (lanes 1 to 3) and DotB K162Q (lanes 4 to 6) ATP binding reactions. Purified protein was incubated with ATP-agarose (lanes 1 and 4), AMP-agarose (lanes 2 and 5), or ATP-agarose in the presence of 10 mM competing free nucleotides (lanes 3 and 6), washed, and eluted. The molecular masses of relevant markers (in kilodaltons) are on the left. Large arrowhead, DotB; small arrowhead, BSA added to reactions to prevent nonspecific binding. (B) Western blot of fractions from binding reactions using L. pneumophila lysate containing wild-type DotB (ΔdotB strain JV918 with DotB complementing clone pJB1153). Whole-cell lysate (lane 1) was incubated with ATP-agarose (lanes 2 and 3), AMP-agarose (lanes 4 and 5), or ATP-agarose in the presence of 10 mM competing free nucleotide (lanes 6 and 7), washed, and eluted. Flowthrough (lanes 2, 4, and 6) and elution (lanes 3, 5, and 7) fractions were collected and subjected to DotB Western blotting. (C) Western blot of fractions from binding reactions using L. pneumophila lysate containing DotB K162Q (ΔdotB strain JV918 with DotB K162Q clone pJB1568). Whole-cell lysate (lane 1) was incubated with ATP-agarose (lanes 2 and 3), AMP-agarose (lanes 4 and 5), or ATP-agarose in the presence of 10 mM competing free nucleotides (lanes 6 and 7), washed, and eluted. Flowthrough (lanes 2, 4, and 6) and elution (lanes 3, 5, and 7) fractions were collected and subjected to DotB Western blotting. The molecular masses of relevant markers (in kilodaltons) are on the left.