Figure 5.
Pol V Mediates Nucleosome Positioning
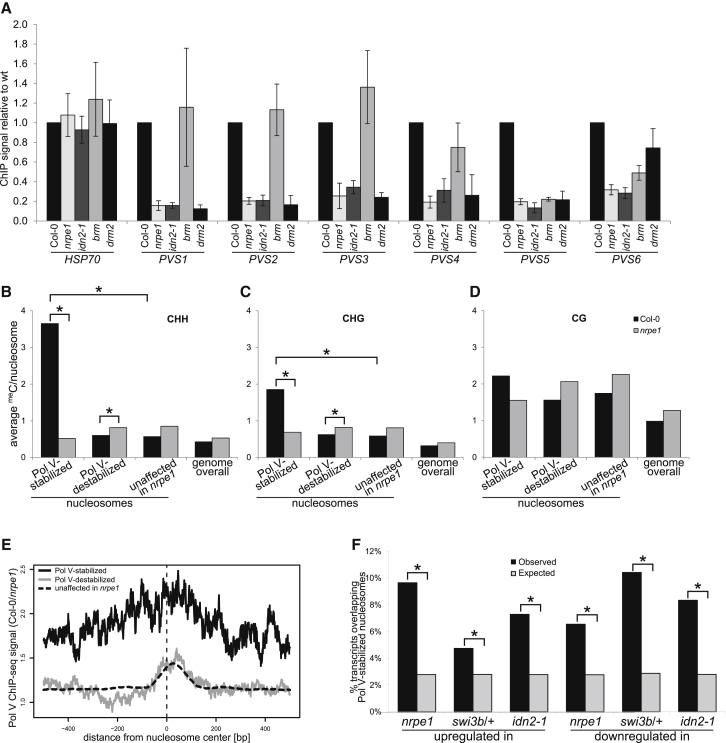
(A) Validation of Pol V-stabilized nucleosomes. Nucleosomes identified with genome-wide assays (Figure S5B) were assayed in seedlings and in case of brm in mature leaves with MNase digestion followed by H3 ChIP and real-time PCR. ChIP signal values were normalized to ACTIN2 and Col-0 wild-type. HSP70 is a negative control (Kumar and Wigge, 2010). Bars show averages and SD from three biological repeats.
(B–D) Nucleosomes stabilized by Pol V are enriched in Pol V-dependent DNA methylation. Published genome-wide DNA methylation data sets from Col-0 wild-type and nrpe1 mutant (Zhong et al., 2012) were used to calculate average DNA methylation levels on nucleosomes identified by MNase-seq and H3 ChIP-seq. Pol V-stabilized and Pol V-destabilized nucleosomes were compared to nucleosomes unaffected in nrpe1 and to the entire genome (genome overall). DNA methylation levels were independently calculated in CHH (B), CHG (C), and CG (D) contexts. Asterisks indicate significant enrichment (see the main text).
(E) Pol V-stabilized nucleosomes are enriched in Pol V binding. A published Pol V ChIP-seq data set (Wierzbicki et al., 2012) was used to calculate profiles of Pol V binding around the centers of nucleosomes identified using MNase-seq and H3 ChIP-seq, which are Pol V-stabilized, Pol V-destabilized, or unaffected in nrpe1.
(F) Pol V-stabilized nucleosomes are enriched on genes upregulated or downregulated in nrpe1 and swi3b/+ mutants. Nucleosomes identified with MNase-seq as Pol V-stabilized were overlapped with genes identified with RNA-seq as upregulated or downregulated in nrpe1 or swi3b. Permutations of gene sets were overlapped in parallel to calculate enrichment. Asterisks indicate significant enrichment (p < 0.02).