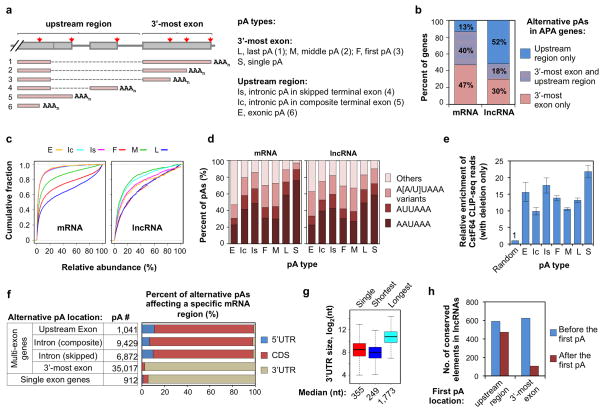
Figure 4. APA of mouse mRNA and lncRNA genes.
(a) Schematic of pA types. The full and short names for different pA types are indicated. The type number in parenthesis corresponds to the isoform number shown in the graph. Dotted lines indicate splicing. (b) Distribution of alternative pAs in different regions of mRNA or lncRNA genes. The p-value for the difference in distribution between mRNA and lnRNA genes is 0 (Chi-squared test). (c) Relative abundance of APA isoforms using different types of pA. The cumulative fraction curve is based on all genes analyzed in this study and on all samples combined. (d) Frequency of various PAS types for different types of pAs in mRNA and lncRNA genes. (e) Enrichment of CstF64 CLIP-seq reads around different types of pAs relative to randomly selected gene regions. Error bars represent the 90% confidence interval derived from bootstrapping (1,000 x) of data. (f) mRNA regions affected by alternative pAs. pAs were grouped based on gene type (multi-exon or single exon) and pA location. mRNA regions were separated into 5′UTR, CDS and 3′UTR. For intronic pAs, the mRNA region affected was defined by the exon immediately upstream of the pA. (g) Distribution of 3′UTR length for genes without alternative pAs in the 3′UTR (single), and genes with APA in the 3′UTR. For the latter, the shortest and longest isoforms were used for analysis. (h) APA regulates conserved elements in lncRNAs. Conserved elements are based on 30 mammalian species (see Methods for detail). The numbers of the conserved elements upstream or downstream of the first pA were calculated. In total, 599 and 391 lncRNA genes have the first pA in the upstream region and 3′-most exon, respectively. Only isoforms with relative expression level > 20% were analyzed.