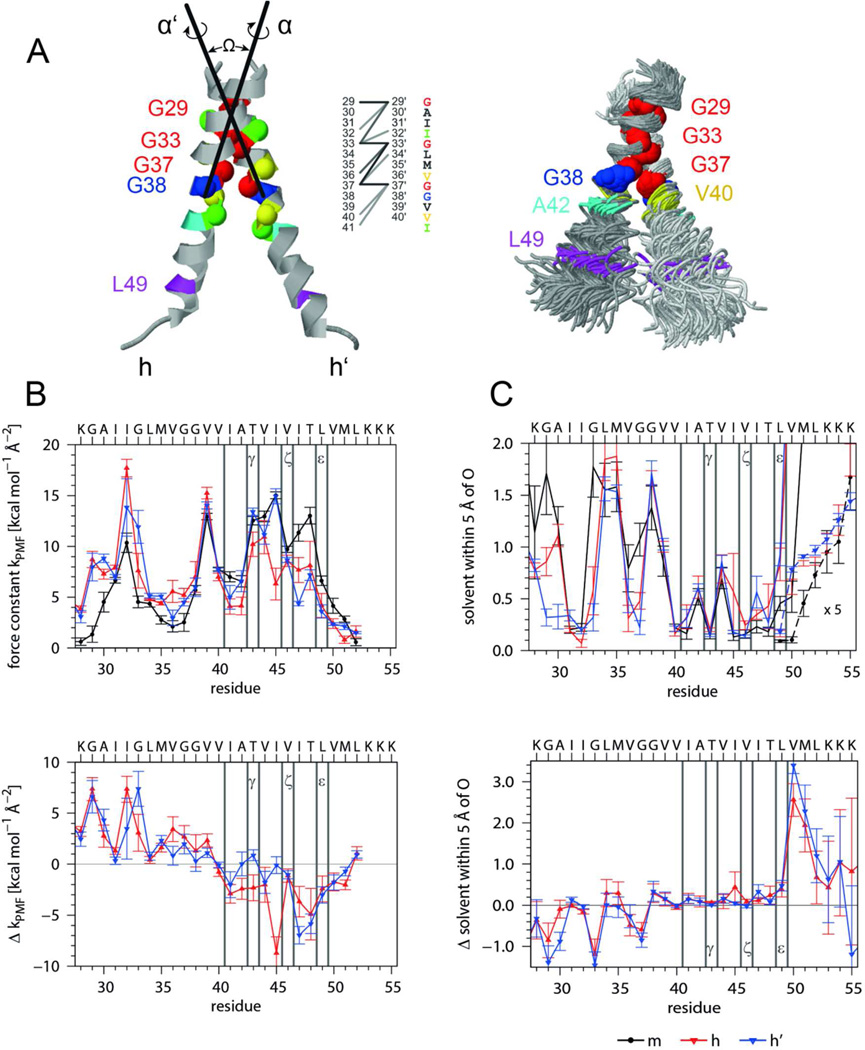
Figure 6. Interhelical interactions, backbone dynamics, and solvation of the A28-55 dimer characterized by MD simulation in isotropic solvation.
A) Average structure and helix-helix contacts (left graph). On average there are 15 contacts formed at any frame of the trajectory as defined by Cα-Cα' distances < 0.6 nm with >50% occupancy (dark, d < 0.5 nm; medium, 0.5 < d/nm < 0.55; light, 0.55 < d/nm < 0.6). The superposition (right graph) shows 100 snapshots taken every ns oriented by a rigid body fit to the Cα atoms of residues 29–37 of the average structure. As compared to the monomer (Fig. 5 A) the bending anisotropy of the helices is reduced. The average interface is slightly asymmetric since interfacial residues of one subunit also contact residues closer to the N-terminus of the partner subunit in addition to their equivalent counterparts. The simulation produced two slightly different dimer populations. The main population (65%) has a crossing angle −50° < Ω < −30° (most probable: −45°) and a center of mass (COM-COM) distance of 0.71 ± 0.03 nm. The helices are oriented (rotation angles α/α’ = −15±15°/ −130±15°) such that the G29xxxG33xxxG37 motifs point toward each other and optimize contact. The minor population with −80° < Ω < −50° (most probable: −60°) has a larger COM-COM distance up to 0.95 nm and the second helix is rotated by 270 ° (α/α’ = −30±10°/ +150±10°) diminishing the contact at G29. B) Helix dynamics characterized by force constants of intra-helical H-bonds extending from carbonyl oxygens at residue i to the amide of residue i+4 or i+3 (upper panel, m = monomeric helix, h, h’ = helices in the dimer) as well as by the differences between mean dimer and monomer values (lower panel). C) Solvation of helices as determined by the numbers of solvent (water or TFE) oxygens within 0.5 nm of the backbone carbonyl oxygens (upper panel, values at V50 to K55 are multiplied by a factor of 5) and differences between mean dimer and monomer values (lower panel). Note that the variation of H-bond strengths of the individual helices is related to the extent of solvation. Error bars indicate standard deviations calculated from 30 ns block averages. Vertical lines denote cleavage sites.