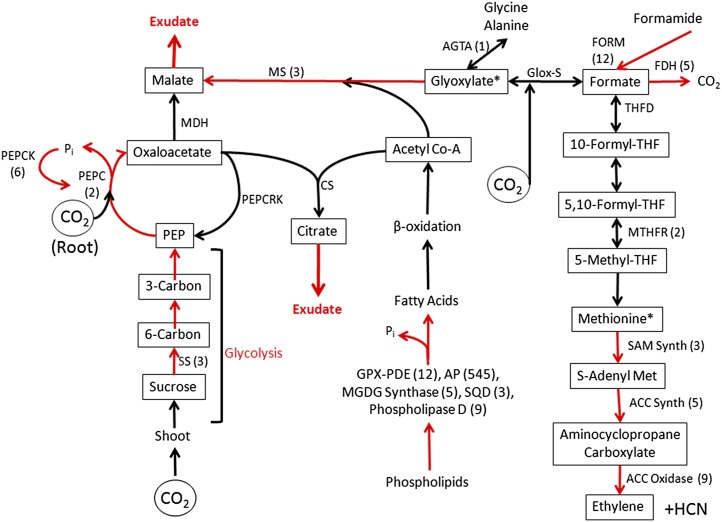
Figure 2.
Adjustments in root metabolism promote acclimation to Pi deficiency. Shown are modifications in white lupin cluster root metabolism that facilitate acclimation to Pi deficiency as evidenced by transcript expression. Increased expression in PdCR was confirmed by qPCR as indicated by fold change in parentheses. Red arrows represent genes known to be up-regulated due to Pi deficiency. Increased Suc metabolism via glycolysis and organic acid production provide carbon for malate and citrate exudation into the rhizosphere. Organic acids lost through exudation are replenished through anaplerotic pathways involving phosphoenolpyruvate carboxylase (PEPC) and a glyoxylate-like cycle malate synthase (MS). One-carbon metabolism is enhanced through a formate and THF pathway. The THF pathway contributes to Met and ethylene production. Increased expression of transcripts involved in phospholipid degradation releases Pi for recycling and carbon for acetyl-CoA synthesis. Acetyl-CoA and glyoxylate provide carbon for malate synthesis through malate synthase. Formate may be carboxylated to glyoxylate by a putative glyoxylate synthase (Glox-S). Additional abbreviations are as follows: Suc synthase (SS); phosphoenolpyruvate carboxylase kinase (PEPCK); phosphoenolpyruvate carboxylase carboxykinase (PEPCRK); malate dehydrogenase (MDH); citrate synthase (CS); formamidase (FORM); formate dehydrogenase (FDH); Ala glyoxylate transaminase (AGTA); THF deformylase (THFD); methylene-THF reductase (MTHFR); S-adenosyl-Met synthase (SAM Synth); aminocyclopropane synthase (ACC Synth); aminocyclopropane oxidase (ACC Oxidase); phospholipase A (PLA1); glycerophosphodiester-phosphodiesterase (GPX-PDE); acid phosphatase (AP); monogalactosyldiacylglycerol synthase (MGDG synthase); sulfoquinovosyltransferase (SQD).