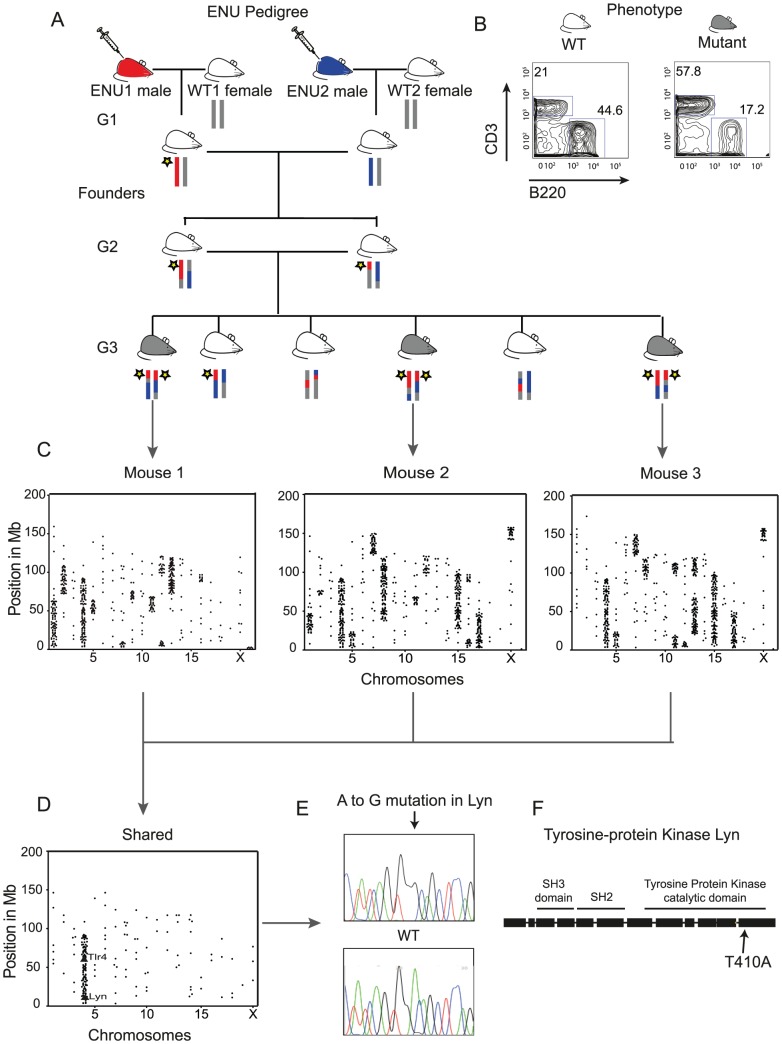
Figure 1. Whole-Genome Sequencing Identifies the IBD Homozygous Region and Causative ENU Mutation.
(A) The structure of an ENU pedigree: two ENU treated males paired with WT B6 females generate founder G1 mice for the ENU16CH17a pedigree, and G3 mice exhibiting the phenotype are selected for WGS. Thus mice within the pedigree carry 4 possible haplotypes, ENU1, ENU2, WT1 and WT2. A yellow star illustrates the segregation of a causative variant. (B) Mice homozygous for the mutation exhibit B cell lymphopenia (here gating on blood lymphocytes). (C) Plots of homozygous filtered variants show the haplotype blocks across the chromosomes of each sequenced mouse. (D) Shared homozygous variants seen in all 3 sequenced mice cluster in an IBD region on Chromosome 4, containing exonic mutations in two genes, Lyn and Tlr4. (E) Confirmation of the Lyn A to G transition by Sanger sequencing. (F) The mutation lies in exon 12 within the catalytic domain.