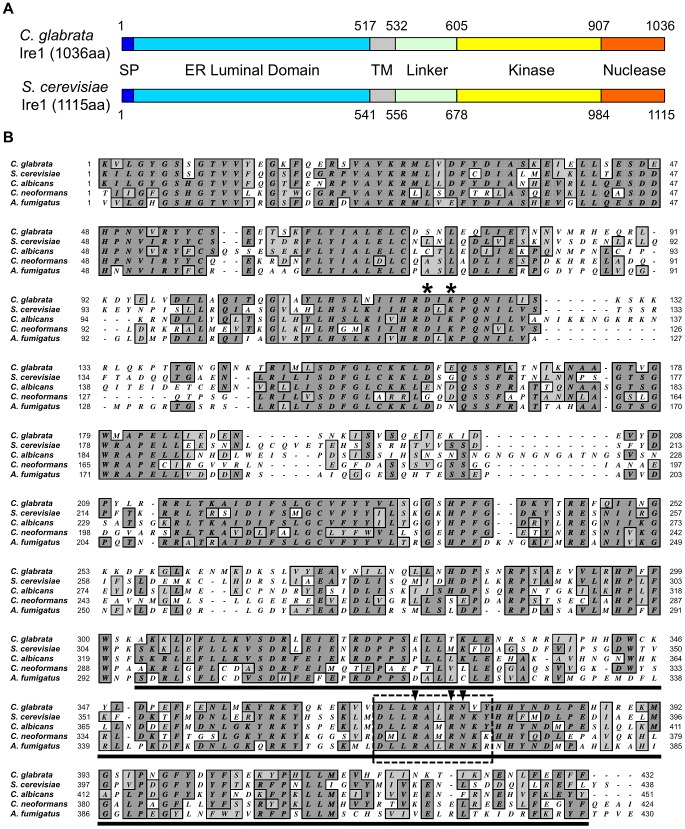
Figure 1. Sequence analysis of C. glabrata Ire1.
(A) Schematic representation of the conserved domain structure of C. glabrata and S. cerevisiae Ire1. Abbreviations: SP, signal peptide; and TM, transmembrane. (B) Sequence alignment of the kinase and nuclease domains of fungal Ire1 orthologs. The asterisk indicates the conserved two catalytic residues in the nucleotide-binding pocket of Ire1 kinase (D797 and K799 in S. cerevisiae and D723 and K725 in C. glabrata). The predicted nuclease domain is underlined. Ten residues (boxed with dotted line) including the highly conserved three-nuclease active sites (arrowheads) are deleted in C. glabrata IRE1-ND. GenBank accession number: Candida glabrata Ire1, XP_446111; Saccharomyces cerevisiae Ire1, NP_011946; Candida albicans Ire1, XP_717532; Cryptococcus neoformans Ire1, XP_568837; and Aspergillus fumigatus IreA, AEQ59230.