FIGURE 4.
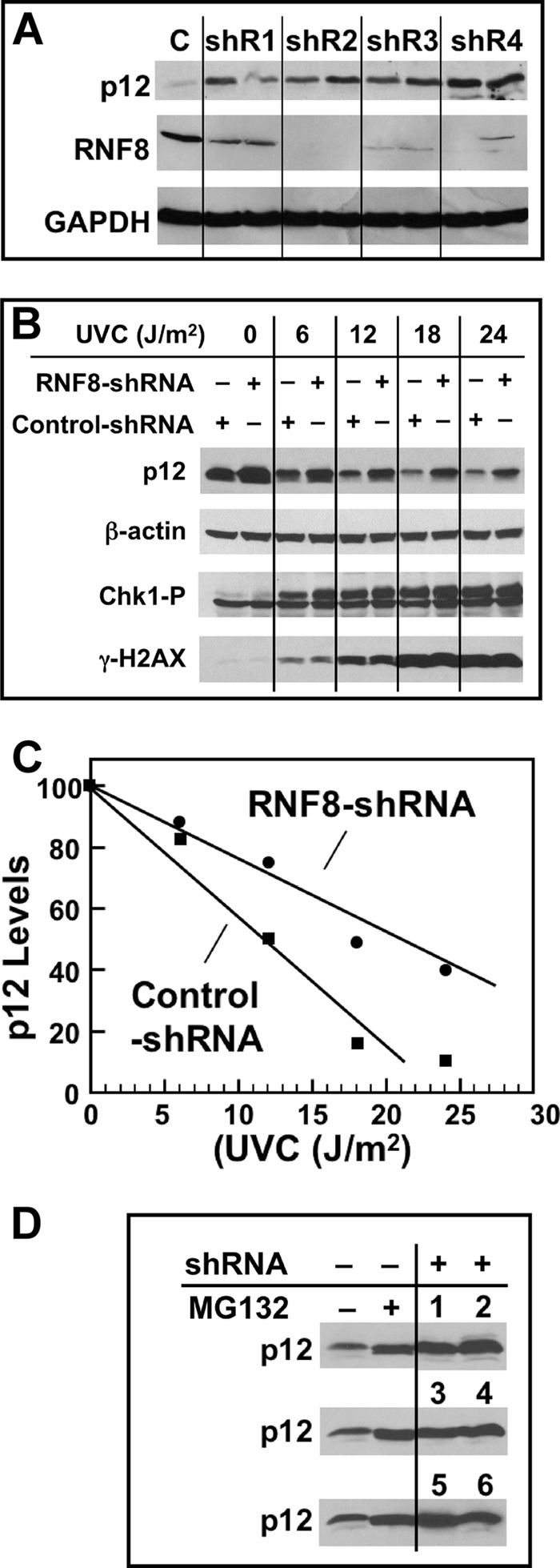
Effects of RNF8 depletion on p12 degradation by UV. A, four different shRNAs (shR1–shR4) and a control shRNA (C) were used to produce respective stable A549 cells (see ”Experimental Procedures“). Each sample was treated in duplicate with UVC (20 J/m2) and Western blotted for RNF8 and p12 after 4 h. GAPDH was used as a loading control. B, UV dose dependence of p12 degradation in RNF8-depleted A549 cells; shR2 was used in these and the following experiments. Cell cultures were treated with UVC (0, 6, 12, 18, and 24 J/m2) and analyzed 4 h later by Western blotting for p12, Chk1-p, γH2AX, and β-actin (loading control). C, the blots for p12 were quantified by densitometry and normalized against the loading control. The relative levels of p12 were then plotted against UVC dose. D, comparison of p12 levels in unperturbed RNF8 knockdown cells. The two left lanes show p12 levels in control shRNA-treated cells in the absence and presence of the proteasome inhibitor MG132 (10 μm), which was added 30 min prior to analysis. The two right lanes with the three rows of numbers show p12 levels in six individual shR2 RNF8 clonal isolates without MG132.
