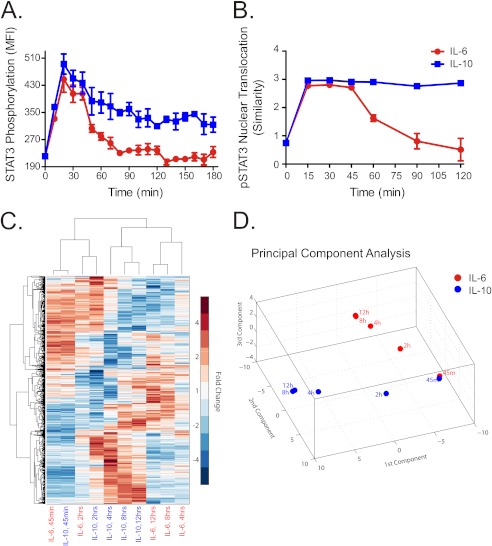
FIGURE 5.
Nuclear translocation of STAT3 and dynamic genome-wide transcriptional responses to IL-6 and IL-10. A, time course of STAT3 tyrosine 705 phosphorylation in IL-6–treated (red) and IL-10–treated (blue) DCs, measured by flow cytometry. MFI is median fluorescence intensity. Error bars are S.D. of MFI of three samples from the same blood donor. B, time course of the nuclear translocation of phosphorylated STAT3 (pSTAT3) in response to IL-6 (red) or IL-10 (blue). For each cell, the pixel-by-pixel correlation, ρ, between the pSTAT3 and nuclear fluorescence intensity images was determined, and similarity was calculated (see “Experimental Procedures”). The median similarity ± S.D. (error bars) of all cells in three samples are shown. C, hierarchical clustering by sample treatment/duration and by genes was performed using the median -fold change in expression relative to untreated cells. Rows represent differentially expressed genes, and columns represent different sample treatments and durations. D, principal component analysis of differentially expressed genes in response to IL-6 (red) or IL-10 (blue) stimulation. The three components plotted account for ∼90% of the total variance.