Background: The GPR-Gαi complex has diverse functional roles, but regulatory mechanisms are not defined.
Results: The GPR-Gαi complex is regulated by Ric-8A but not by increased expression of AGS1 or GIV/Girdin.
Conclusion: The GPR-Gαi complex is differentially regulated by specific guanine nucleotide exchange factors.
Significance: The GPR proteins, Gαi1 and Ric-8A, exhibit dynamic interactions in the cell that influence their subcellular localization and regulate complex formation.
Keywords: Cell Biology, G-protein-coupled Receptors, G-proteins, Guanine Nucleotide Exchange Factor (GEF), Signal Transduction, BRET, G-protein Signaling Modulator, G-protein Regulatory Motif
Abstract
Group II activators of G-protein signaling (AGS) serve as binding partners for Gαi/o/t via one or more G-protein regulatory (GPR) motifs. GPR-Gα signaling modules may be differentially regulated by cell surface receptors or by different nonreceptor guanine nucleotide exchange factors. We determined the effect of the nonreceptor guanine nucleotide exchange factors AGS1, GIV/Girdin, and Ric-8A on the interaction of two distinct GPR proteins, AGS3 and AGS4, with Gαil in the intact cell by bioluminescence resonance energy transfer (BRET) in human embryonic kidney 293 cells. AGS3-Rluc-Gαi1-YFP and AGS4-Rluc-Gαi1-YFP BRET were regulated by Ric-8A but not by Gα-interacting vesicle-associated protein (GIV) or AGS1. The Ric-8A regulation was biphasic and dependent upon the amount of Ric-8A and Gαi1-YFP. The inhibitory regulation of GPR-Gαi1 BRET by Ric-8A was blocked by pertussis toxin. The enhancement of GPR-Gαi1 BRET observed with Ric-8A was further augmented by pertussis toxin treatment. The regulation of GPR-Gαi interaction by Ric-8A was not altered by RGS4. AGS3-Rluc-Gαi1-YFP and AGS4-Rluc-G-Gαi1-YFP BRET were observed in both pellet and supernatant subcellular fractions and were regulated by Ric-8A in both fractions. The regulation of the GPR-Gαi1 complex by Ric-8A, as well as the ability of Ric-8A to restore Gα expression in Ric8A−/− mouse embryonic stem cells, involved two helical domains at the carboxyl terminus of Ric-8A. These data indicate a dynamic interaction between GPR proteins, Gαi1 and Ric-8A, in the cell that influences subcellular localization of the three proteins and regulates complex formation.
Introduction
Activators of G-protein signaling (AGS)4 proteins were identified in a functional screen for cDNAs that activated G-protein signaling in the absence of a seven-membrane span receptor (1, 2). The discovery of such regulatory mechanisms led to the concept that Gα and Gβγ regulate intracellular events distinct from their role as transducers for cell surface seven-transmembrane span receptors (2–15). Such regulatory mechanisms include a panel of accessory proteins that may serve as binding partners for Gα and Gβγ independent of the classical heterotrimer Gαβγ. AGS proteins may also influence Gα-Gβγ interaction and modulate guanine nucleotide binding or hydrolysis by Gα (1, 2, 10, 16). AGS proteins are involved in a wide spectrum of biological effects (17–29) providing attractive mechanisms for tissues to respond and adapt to physiological and pathological challenges.
One group of accessory proteins of particular interest is defined by the group II AGS proteins (16), all of which contain one or more G-protein regulatory (GPR) motifs, also known as GoLoco or LGN motifs (30, 31), that stabilize the GDP-bound conformation of Gα serving as alternative binding partners for Gα-GDP (Gαi, Gαt, and/or Gαo) free of Gβγ. Group II AGS proteins (AGS3 (GPSM1), LGN (GPSM2/AGS5), AGS4 (GPSM3), RGS12 (AGS6), Rap1Gap (transcript variant 1), RGS14, and PCP2/L7 (GPSM4)) provide 1–4 docking sites for Gα and form complexes with subpopulations of Gαi class subunits in the cell. There are three types of group II AGS proteins. One group includes AGS3 and LGN (AGS5), both of which have a tetratricopeptide repeat region that is separated from a series of four GPR motifs by an extended linker region. The second group of proteins (AGS3-Short, AGS4, and Pcp2/L7) contains three GPR motifs without any other obvious protein interaction domains. The third group of proteins (RGS12, RGS14, and Rap1GAP) has one GPR motif plus other defined domains that act to accelerate Gα-GTP hydrolysis (16).
The GPR-Gα signaling module may also exist in different subcellular compartments where it is differentially regulated and involved in discrete biological events such as the control of asymmetric cell division. One central question is what are the mechanisms that regulate interactions between GPR-containing proteins and their G-protein partners? Such signals may involve regulated positioning of the proteins within the cell, second messengers, and/or guanine nucleotide exchange factors (GEFs) that act on the GPR-Gαi complex (2, 3, 27, 32–41), and these questions must be addressed in the intact cell. We recently reported regulation of the AGS3-Gαi and AGS4-Gαi signaling cassettes by a cell surface seven-transmembrane span receptor and suggested that the GPR-Gαi module was one component of a larger signaling complex at the cell cortex (42, 43).
GPR-Gαi signaling modules are also regulated by nonreceptor GEFs and may operate independently of the classical membrane receptor-Gαβγ signaling system. Such nonreceptor GEFs include the group I AGS protein AGS1 (dexamethasone-induced Ras-related protein or Rasd1), which interacts with Gαi1 and Gαo and increases GTPγ35S binding to purified heterotrimeric brain G-protein and purified Gαi and Gαo (1, 44). AGS1 is also reported to interact with Gβ (45). GIV/Girdin (coiled-coil domain containing 88A) interacts with Gαi3 and acts as a GEF for AGS3-Gαi3 (36). The Gα-interacting vesicle-associated protein (GIV) carboxyl terminus (GIV(1660–1870)) increased the apparent Gαi guanine nucleotide exchange rate (6). The nonreceptor GEF Ric-8A (resistance to inhibitors of cholinesterase 8A homolog, synembryn-A) directly regulated purified AGS3-GPR-Gαi1, AGS5/LGN-Gαi1, and RGS14-Gαi1 complexes (39–41, 46). Thus, GPR-Gα signaling modules may be regulated by nonreceptor GEFs and operate independently of the classical membrane receptor-Gαβγ signaling system. As is the case for G-protein-coupled receptors coupled to Gαβγ, various members of the family of regulators of G-protein signaling (RGS) may also accelerate guanine nucleotide hydrolysis following nonreceptor GEF-mediated generation of Gα-GTP from GPR-Gα-GDP.
AGS1 is a Ras-related protein that regulates the ERK1/2 signaling pathway and cell growth (44, 47–49). Loss of AGS1 is associated with breast cancer, and alterations in AGS1 expression are observed in prostate cancer, renal cell carcinoma, dexamethasone-resistant multiple myeloma, and oligodendroglial tumors in response to chemotherapy (50–54). GIV/Girdin may process signals from the epidermal growth factor receptor to regulate autophagy and metastasis (36, 55). GIV/Girdin expression is increased in gastrointestinal cancers (56). Among the nonreceptor GEFs studied to date, Ric-8A is perhaps the best characterized biochemically in terms of its interaction with G-protein subunits and its action as a GEF. Genetically based approaches in the model organisms Drosophila melanogaster and Caenorhabditis elegans indicate a role for the Ric-8 ortholog in asymmetric cell division during early development, which involves Gα and GPR proteins (57–64). A similar functional role for Ric-8A was recently reported in mammalian systems (65). In addition to the apparent role of Ric-8A as a molecular chaperone for Gα (66), Ric-8A may also play a variety of roles in signal processing.
We recently developed an experimental approach to monitor the interaction of AGS3 and AGS4 with Gαi in the intact cell by bioluminescence resonance energy transfer (BRET) following expression of proteins tagged with Renilla luciferase (Rluc) and yellow fluorescent protein (YFP) (42, 43, 67–69). Co-expression of AGS3-Rluc or AGS4-Rluc with Gαi1-YFP generates robust, specific BRET that results from binding of multiple Gαi1 subunits to the GPR domains of AGS3 and AGS4. The interaction of Gαi1-YFP with AGS3-Rluc or AGS4-Rluc stabilized the GPR protein at the cell cortex where the GPR-Gαi1 module was regulated by activation of cell surface receptors (42, 43). We used this system to determine the effect of nonreceptor GEFs on the interaction of Gαil with two different types of GPR proteins, AGS3 and AGS4.
The functional role of AGS4 has not been determined, but it is of particular interest due to its relatively restricted expression to immune system tissues and the role of G-protein systems in the immune cell response. AGS3 has multiple functional roles in asymmetric cell division, neuronal plasticity and addiction, autophagy, membrane protein trafficking, polycystic kidney disease, cardiovascular regulation and metabolism (3, 17–20, 22, 24, 28, 29, 70–73). LGN (AGS5/GPSM2), which is closely related to AGS3, also plays important functional roles in asymmetric cell division and morphogenesis and was recently identified as a responsible gene for certain types of nonsyndromic hearing loss as well as for the brain malformations and hearing loss in Chudley-McCullough syndrome (26, 74).
EXPERIMENTAL PROCEDURES
Materials
AGS1 (RASD1) antisera were generated as described previously (49). Anti-His6 antibody was obtained from Pharmingen. Ric-8A antisera was described previously (39). Ric-8A (NP_001093990.1), Ric-8A(Met1–Asn492), and Ric-8A(Met1–Asn453) cDNA were cloned into pcDNA3.1 as described previously (12). Full-length GIV/Girdin (NP_ 001129069.1 Girdin isoform 1) was kindly provided by Drs. Mikel Garcia-Marcos (Boston University, Boston) and Marilyn Farquhar (University of California, San Diego). Full-length GIV was used as template in PCRs using Takara Taq (Fisher) to generate pcDNA3.1/His::GIV(1660–1870). The following oligonucleotides and restriction enzymes were used in the PCR amplification and subsequent digestion: BamHI, GIV(1660–1870) forward primer, 5′-TCG GAT CCA CAC CAT GTC TGA AAC ACT GGA GAG TCG A-3′; EcoRI, GIV(1660–1870) reverse primer 5′-GCA GAA TTC GGA GCT TTG TTG CTC CCT AGA CCT-3′. AGS1 (NP_057168, dexamethasone-induced Ras-related protein 1 isoform 1 proprotein) was cloned into pcDNA3.1/His and provided by Dr. Mary Cismowski (Nationwide Children's Hospital, Columbus, OH). Complete protease inhibitor mixture tablets were purchased from Roche Applied Science and used in accordance with manufacturer's instructions. All other reagents and materials were obtained as described elsewhere (12, 42, 43, 75). Ric-8A−/− mouse embryonic stem (ES) cells were generated and processed for complementation assays as described elsewhere (66).
Cell Culture, Transfection, Immunoblotting, Bioluminescence Resonance Energy Transfer (BRET)
The human epithelial cell line (HEK-293) and neuronal catecholaminergic cell line (CAD) was maintained in Dulbecco's minimal essential medium (high glucose, without phenol red) supplemented with 10% fetal bovine serum, 2 mm glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin. Cells were grown in a humidified incubator in the presence of 5% CO2 at 37 °C. These procedures are described elsewhere in detail (42, 43). Generally, cells were transfected with a fixed amount of phRluc::AGS3 (10 ng) or phRluc::AGS4 (2 ng) and increasing amounts of pcDNA3::Gαi1-YFP without or with varying amounts of pcDNA3::Ric-8A, pcDNA3::AGS1, or pcDNA3::GIV(1660–1870). The total plasmid load was harmonized by including the appropriate amount of pcDNA3 vector. For brevity, plasmid amounts used for transfection are indicated as nanograms in the figures. The plasmid amounts used for BRET measurements result in levels of the individual tagged proteins that are comparable with that of the endogenous protein (43).
Cell Lysis and Fractionation
Cells were split into 6-well tissue culture plates and transfected with phRluc::AGS3 or phRluc::AGS4, pcDNA3::Gαi1-YFP, and/or pcDNA3::Ric8A (42, 43). Forty eight hours later, cells were suspended in BRET buffer (750 μl/well) (42, 43), and the suspensions from six wells were pooled. 300 μl (∼300,000 cells) of the pooled suspension were used for intact cell measurements of fluorescence, luminescence, and BRET, and the remainder was processed for subcellular fractionation. Total fluorescence (excitation, 485 nm; emission, 535 nm) was measured to determine the total cellular levels of Gαi1-YFP. Luciferase substrate coelenterazine H (5 μm final concentration) was then added and luminescence measured at 480 ± 20 nm to determine the level of AGS3-Rluc or AGS4-Rluc. The remaining pooled suspension (4.2 ml) was centrifuged (200 × g, 5 min), and the pellet was lysed in 0.8 ml of hypotonic lysis buffer (5 mm EDTA, 5 mm EGTA, 5 mm Tris-HCl, pH 7.4, and protease inhibitor mixture) with a 26-gauge syringe followed by centrifugation at 10,000 × g for 10 min to obtain crude membrane (pellet) and cytosol (supernatant) fractions. Pellets were resuspended in 300 μl of membrane buffer (50 mm Tris-HCl, pH 7.4, 5 mm MgCl2, 1 mm EDTA, 1 mm EGTA, and protease inhibitor mixture). Pellet (50 μg of protein) and supernatant (50 μg of protein) samples from each group were then mixed with 150 μl of BRET buffer for measurements of fluorescence, luminescence, and BRET as described above.
Data Analysis
Statistical significance for differences involving a single intervention was determined by the Student's t test as noted in figure and table legends. Data involving multiple treatment paradigms were analyzed by analysis of variance and significant differences between groups determined by the Tukey a posteriori test using GraphPad Prism version 4.03 for Windows (GraphPad Software, San Diego).
RESULTS AND DISCUSSION
Ric-8A, GIV/Girdin, and AGS1/RASD1 as Nonreceptor GEFs
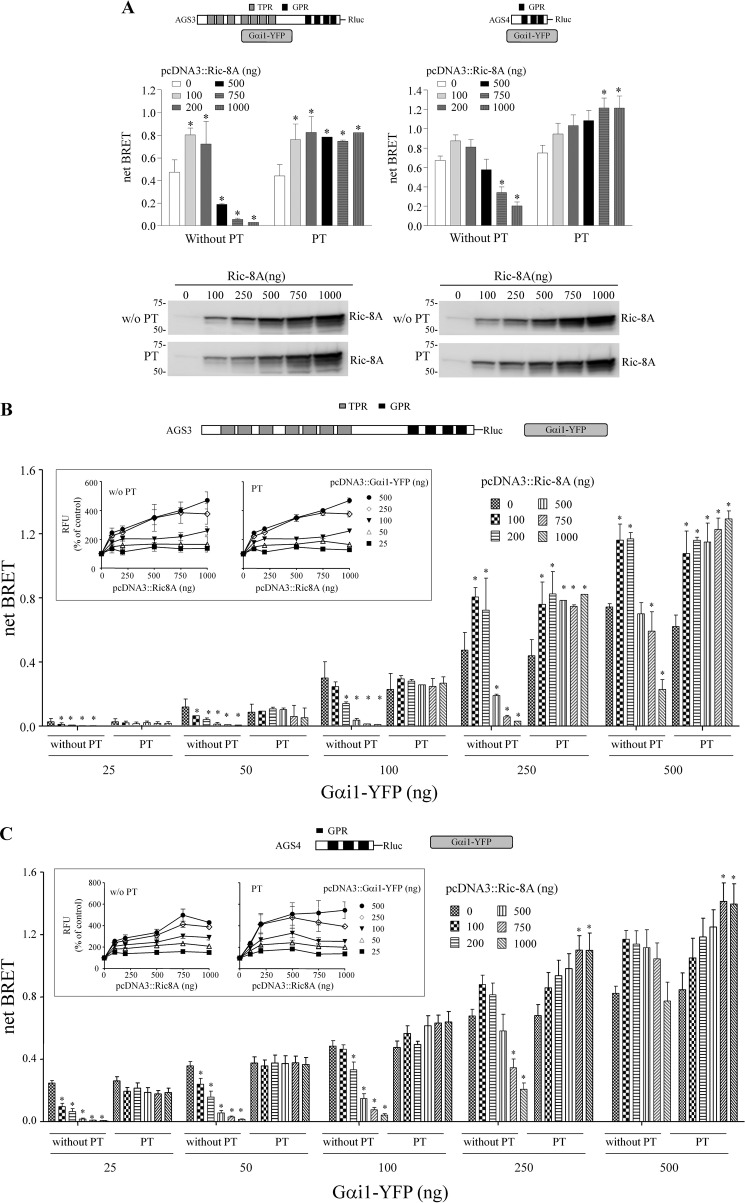
In contrast to cell surface seven-transmembrane span receptors, nonreceptor guanine nucleotide exchange factors for G-proteins, such as Ric-8A, GIV/Girdin, and AGS1/RASD1, are not embedded in the plasma membrane, and they may differ from the receptor in terms of their mechanisms of action and/or the subpopulations of G-proteins that they regulate. We first determined the effects of Ric-8A, AGS1, and GIV(1660–1870) on the GPR-Gαi1 interaction using our previously established BRET platforms for AGS3 and AGS4 (Fig. 1, A and B) (42, 43). AGS3 and AGS4 were selected as representative group II AGS proteins that contain clearly defined regulatory domains in tandem with the GPR domain (AGS3) or primarily consist of the GPR core domain (AGS4). Neither GIV(1660–1870) nor AGS1 altered AGS3-Rluc-Gαi1-YFP or AGS4-Rluc-Gαi-YFP BRET. Full-length GIV/Girdin also had no effect.5
FIGURE 1.
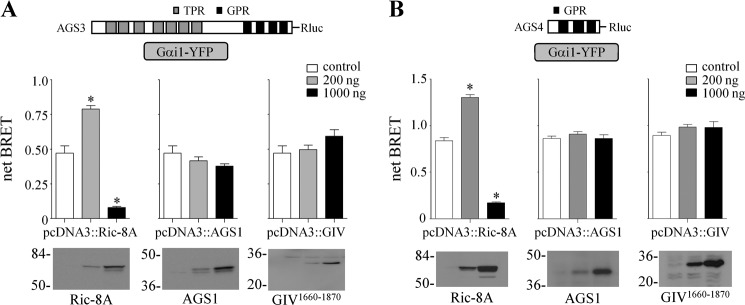
Regulation of AGS3-Rluc-Gαi1-YFP and AGS4-Rluc-Gαi1-YFP BRET by nonreceptor GEFs. AGS3-Rluc (10 ng) (A) and AGS4-Rluc (2 ng) (B) were expressed together with Gαi1-YFP (250 ng) in HEK cells, and BRET was measured as described under “Experimental Procedures.” Ric-8A, AGS1, or GIV(1660–1870) was expressed as indicated. Data are presented as the mean ± S.E. from four experiments with triplicate determinations. *, p < 0.05 compared with control. The relative fluorescent units and relative luciferase units for sample points in A and B are presented in Table 1. Lower panels, Ric-8A, AGS1, and GIV(1660–1870) immunoblots. Ric-8A and AGS1 proteins were detected with affinity-purified anti-Ric-8A and anti-AGS1 antibody, respectively. GIV(1660–1870) was detected with anti-His6 antibody. Each lane contains 50 μg of total protein, and the immunoblot is representative of three separate experiments. The GIV construct (pcDNA3.1/His::GIV(1660–1870)) encoded the carboxyl-terminal region of the protein as described under “Experimental Procedures.”
In contrast, AGS3-Rluc-Gαi1-YFP and AGS4-Rluc-Gαi1-YFP BRET were both regulated by Ric-8A (Fig. 1). The regulation appeared to be biphasic depending upon the level of Ric-8A expression. Ric-8A also increased Gαi1-YFP expression levels (Table 1).
TABLE 1.
Influence of nonreceptor guanine nucleotide exchange factors on the expression levels of AGS3-Rluc, AGS4-Rluc, and Gαi1-YFP
RFU and RLU generated for the data set presented in Fig. 1 A and B, were measured as described under “Experimental Procedures” and expressed as percent of control values obtained in the absence of pcDNA3::Ric-8A transfection. RFU and RLU control values for Fig. 1A were 125,350 ± 27,902 and 383,258 ± 134,332, respectively. RFU and RLU control values for Fig. 1B were 108,727 ± 2,606 and 421,370 ± 27,315, respectively. The GIV construct (pcDNA3.1/His::GIV(1660–1870)) encoded the carboxyl-terminal region of the protein as described under “Experimental Procedures.” Results are expressed as the mean ± S.E. of four independent experiments with triplicate determinations.
| pcDNA3::Ric-8A |
pcDNA3::AGS1 |
pcDNA3::GIV |
|||||
|---|---|---|---|---|---|---|---|
| 200 | 1,000 | 200 | 1,000 | 200 | 1,000 | ||
| ng | ng | ng | |||||
| Fig. 1A | RFU | 225 ± 27 | 504 ± 80a | 100 ± 10 | 99 ± 8 | 127 ± 9 | 167 ± 26 |
| RLU | 91 ± 16 | 239 ± 35 | 68 ± 1 | 69 ± 7 | 84 ± 11 | 114 ± 18 | |
| Fig. 1B | RFU | 329 ± 7a | 478 ± 11a | 90 ± 3 | 84 ± 4 | 105 ± 1 | 146 ± 2a |
| RLU | 102 ± 4 | 135 ± 7a | 85 ± 1 | 82 ± 3 | 110 ± 5 | 116 ± 4 | |
a p < 0.05 compared with control values was determined by analysis of variance as described under “Experimental Procedures.”
The action of Ric-8A was further explored by a series of experiments, including determination of stoichiometric considerations, the effect of pertussis toxin treatment, and the role of nucleotide hydrolysis on the Ric-8A-mediated regulation of AGS3- and AGS4-Gαi BRET. We also examined the effect of Ric-8A on the GPR-Gαi1 complex in subcellular compartments and the subcellular distribution of the binding partners. Finally, we examined the role of the Ric-8A carboxyl-terminal region on the Ric-8A-mediated regulation of endogenous Gα expression and the regulation of the GPR-Gαi1 signaling cassette.
Ric-8A-mediated Regulation of the GPR-Gαi1 Signaling Cassette
For a more complete understanding of the dynamics of the signals generated through resonance energy transfer as a result of protein interaction, the signals were evaluated over a range of acceptor concentrations with a fixed amount of donor. Fig. 2A presents data at one level of Gαi1-YFP expression (250 ng), whereas Fig. 2, B and C, presents data over a range of Gαi1-YFP expression levels. At lower Gαi1-YFP expression levels (25, 50, and 100 ng of pcDNA::Gαi1-YFP), both AGS3-Rluc-Gαi1-YFP and AGS4-Rluc-Gαi1-YFP BRET were reduced by Ric-8A in a manner that was dependent upon the amount of Ric-8A protein (Fig. 2, B and C). At higher Gαi1-YFP expression levels (250 and 500 ng of pcDNA::Gαi1-YFP), the effect of Ric-8A on AGS3-Rluc-Gαi1-YFP and AGS4-Rluc-Gαi1-YFP BRET was biphasic in that the BRET was augmented at the lower expression levels of Ric-8A but inhibited at the higher levels of Ric-8A expression (Fig. 2).6
FIGURE 2.
Regulation of AGS3-Rluc-Gαi1-YFP and AGS4-Rluc-Gαi1-YFP BRET by Ric-8A. A, BRET data presented in A were extracted from the larger, complete datasets in B and C. Lower panels, 1% Nonidet P-40 lysates from HEK cells expressing AGS3-Rluc, AGS4-Rluc, and Gαi1-YFP (250 ng of plasmid) and increasing amounts of Ric-8A as in the upper panel were subjected to SDS-PAGE and immunoblotting with Ric-8A antisera. The immunoblot is representative of two similar experiments. The numbers to the right of the immunoblots correspond to the migration of prestained Bio-Rad protein standards. *, p < 0.05 compared with their control. For AGS3-Rluc (10 ng) (B) and AGS4-Rluc (2 ng) (C), increasing amounts of Gαi1-YFP (25–500 ng) and Ric-8A (as indicated in the figure) were expressed in HEK cells, and BRET was measured as described under “Experimental Procedures.” In some experiments, cells were pretreated with pertussis toxin (100 ng/ml) for 16 h. Data in B and C are presented as the mean ± S.E. from four to five experiments with triplicate determinations. *, p < 0.05 compared with their control. Relative fluorescence units (RFU) were measured for each sample in B and C as described under “Experimental Procedures.” Insets in B and C, data are presented as the percentage of control values (cells expressing only AGS3-Rluc and Gαi1-YFP). Results are expressed as the mean ± S.E. of four to five (B) or three (C) independent experiments with triplicate determinations. Inset for B, RFU values for control cells transfected with 25, 50, 100, 250, and 500 ng of pcDNA3::Gαi1-YFP were 41,729 ± 793, 65,720 ± 2,291, 98,810 ± 2,716, 150,534 ± 6,153, and 178,964 ± 5,871, respectively, and for PT-treated cells were 41,673 ± 2,445, 55,531 ± 3,899, 94,584 ± 6,931, 148,609 ± 5,501 and 160,708 ± 5,583, respectively. RFU values at each level of transfected pcDNA3::Gαi1-YFP, with or without PT, were significantly different (p < 0.05) from the corresponding control value with the exception of the RFU value for pcDNA3::Gαi1-YFP (25 ng), pcDNA3::Ric-8A (200 ng). C, RFU values for control cells transfected with 25, 50, 100, 250, and 500 ng of pcDNA3::Gαi1-YFP were 37,816 ± 1559, 42,803 ± 1,628, 55,727 ± 1,858, 75,420 ± 2,836 and 93,555 ± 5,158, respectively, and for PT-treated cells were 40,313 ± 2,031, 47,200 ± 3,208, 62,215 ± 4,841, 83,990 ± 6,741, and 111,044 ± 7,388, respectively. *, p < 0.05 compared with their control. RFU values at each level of transfected pcDNA3::Gαi1-YFP for PT-treated cells were significantly different (p < 0.05) from the corresponding control value with the exception of the RFU values the following samples: pcDNA3::Gαi1-YFP (25 ng), pcDNA3::Ric-8A (100, 750, and 1,000 ng); pcDNA3::Gαi1-YFP (50, 100, 250, and 500 ng), pcDNA3::Ric-8A (100 ng); pcDNA3::Gαi1-YFP (500 ng), pcDNA3::Ric-8A (200 ng).
As noted earlier, Ric-8A increased Gαi-YFP levels in the cell, and this effect was also dependent upon the expression level of Gαi-YFP and Ric-8A (Fig. 2, B, and C, insets). These data suggest that the effect of Ric-8A on AGS3-Rluc-Gαi1-YFP and AGS4-Rluc-Gαi1-YFP BRET reflects a balance of the inhibitory effect of Ric-8A on the GPR-Gαi1 complex, and the augmentation of BRET as the overall level of Gαi1 is increased by Ric-8A co-expression (59, 60, 64, 75).
We then asked if the biphasic regulation of the GPR-Gαi1 complex by Ric-8A was also observed in other cell types. Similar overall results were obtained in the neuronal catecholaminergic cell line (CAD) indicating that the regulatory mechanisms were not restricted to a specific cell type (Fig. 3).
FIGURE 3.
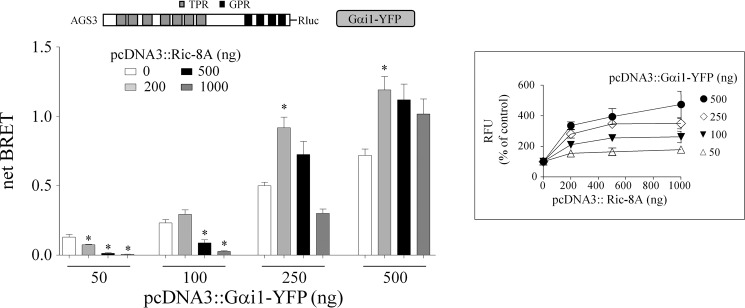
Regulation of AGS3-Rluc-Gαi1-YFP BRET by Ric-8A in the neuronal catecholaminergic cell line (CAD). AGS3-Rluc (10 ng) was expressed together with Gαi1-YFP and Ric-8A as indicated, and BRET was measured as described under “Experimental Procedures.” Data are presented as the mean ± S.E. from three experiments with triplicate determinations. *, p < 0.05 compared with their control. Inset, data are presented as the percentage of control values (cells expressing only AGS3-Rluc and Gαi1-YFP). Results are expressed as the mean ± S.E. of four independent experiments with triplicate determinations. RFU values for control cells transfected with 10 ng of phRluc::AGS3 and 50, 100, 250, and 500 ng of pcDNA3::Gαi1-YFP were 21,969 ± 2,231, 24,097 ± 1,598, 35,383 ± 2,472, and 49,359 ± 4,926, respectively. RFU values at each level of transfected pcDNA3::Gαi1-YFP were significantly different (p < 0.05) from the corresponding control value with the exception of the RFU values for pcDNA3-Gαi1-YFP (50 ng), pcDNA3::Ric-8A (200 and 500 ng).
AGS4 and AGS3 define two different classes of GPR proteins. AGS4 (160 amino acids) contains three GPR motifs without any other defined protein interaction or regulatory motif. In contrast, AGS3 (650 amino acids) contains four GPR motifs and an amino-terminal domain containing seven tetratricopeptide repeats. A third class of GPR proteins consists of RGS12, RGS14, and Rap1Gap. All three classes of GPR proteins are apparently regulated by Ric-8A (Fig. 2) (39–41, 46). Although it is difficult to make direct comparisons, the level of resonance energy transfer exhibited by AGS4-Rluc-Gαi1-YFP was greater than that observed for AGS3-Rluc-Gαi1-YFP, despite apparently similar amounts of protein as reflected by the levels of luciferase activity and fluorescence. These data suggest that the tetratricopeptide repeat domain may modulate the interaction of the GPR motifs with Gαi (35, 42).
Ric-8A-mediated Regulation of the GPR-Gαi1 Signaling Cassette: Effect of Pertussis Toxin Treatment and the GTPase-accelerating Protein RGS4
Pertussis toxin (PT) pretreatment, which interferes with receptor coupling to Gαβγ and Ric-8A-mediated regulation of Gαi, does not inhibit the interaction of GPR proteins with Gαi1 (42, 43, 65). We therefore asked if PT pretreatment influenced the effect of Ric-8A on the GPR-Gαi1 signaling cassette. PT ADP-ribosylates a cysteine residue four amino acids from the carboxyl terminus of Gαi1. PT pretreatment had no effect or slightly increased AGS3-Rluc-Gαi1-YFP and AGS4-Rluc-Gαi1-YFP BRET (42, 43). The inhibitory effect of Ric-8A on AGS3-Rluc-Gαi1-YFP and AGS4-Rluc-Gαi1-YFP BRET was completely blocked by PT pretreatment (Fig. 2).6 The magnitude of AGS3-Rluc-Gαi1-YFP and AGS4-Rluc-Gαi1-YFP BRET observed upon co-expression of Ric-8A was increased following PT treatment (Fig. 2), likely reflecting the increased levels of Gαi1-YFP observed upon expression of Ric-8A and the elimination of any Ric-8A-mediated GEF activity.6 Thus, the magnitude of the increased AGS3-Rluc-Gαi1-YFP and AGS4-Rluc-Gαi1-YFP BRET observed after PT treatment of the cells likely correlates with the magnitude of the inhibitory action of Ric-8A on AGS3-Rluc-Gαi1-YFP and AGS4-Rluc-Gαi1-YFP BRET at the higher concentrations of Gαi1-YFP where the effect of Ric-8A is biphasic.
PT treatment did not prevent the increase in Gαi1-YFP protein observed upon co-expression of Ric-8A (Fig. 2, B and C), which may reflect an action of Ric-8A that occurs before PT treatment and/or the presence of a population of Gαi-YFP that is not an effective substrate for PT and is stabilized by interaction with Ric-8A. However, the Ric-8A-mediated increase in Gαi1-YFP protein was also observed when cells were treated with PT prior to transfection,7 which suggests that Ric-8A interacts with Gαi1-YFP before it becomes an effective substrate for ADP-ribosylation by PT (76). These data with the Ric-8A-mediated enhancement of Gαi1-YFP levels appear to delineate two seemingly independent functions of Ric-8A, one as a GEF for Gαi/o/q subunits and the other as a Gα biosynthetic factor and/or chaperone (66), with the former but not the latter blocked by PT treatment. A trend of minimally increased luciferase activity was also observed with increasing expression of Ric-8A and Gαi1, likely reflecting increased expression of AGS3-Rluc and AGS4-Rluc.8 Notably, this trend was not observed after pertussis toxin treatment suggesting that it was dependent upon the ability of Ric-8A to promote guanine nucleotide exchange.8
The loss of Ric-8A-mediated regulation of the GPR-Gαi1 signaling cassette after PT treatment indirectly suggests that the Gαi1-YFP complexed with AGS3-Rluc or AGS4-Rluc was ADP-ribosylated, as AGS3-Rluc-Gαi1-YFP and AGS4-Rluc-Gαi1-YFP BRET were not altered by Ric-8A after PT pretreatment of the cells. This point is of particular interest, as it would suggest that the GPR-Gαi1 complex is a substrate for PT. Certainly Gαi alone is not an effective substrate for pertussis toxin, but it is ADP-ribosylated by pertussis toxin when it is complexed with Gβγ (76). An analogous situation may exist for Gαi complexed with a GPR motif. Alternatively, Gαi may be ADP-ribosylated by PT when it is complexed with Gβγ and then ADP-ribosylated Gαi is transferred to a GPR protein and such a GPR-Gαi1 complex would not be a substrate for Ric-8A GEF activity. Cellular responses elicited through seven-transmembrane receptors that are blocked by PT pretreatment are categorized as coupling to a subset of heterotrimeric G-proteins. However, as PT treatment also prevents receptor and Ric-8A mediated regulation of the GPR-Gαi1 signaling module, it is plausible that PT-sensitive cellular effects of receptor activation may involve both Gαβγ and GPR-Gα complexes (65).
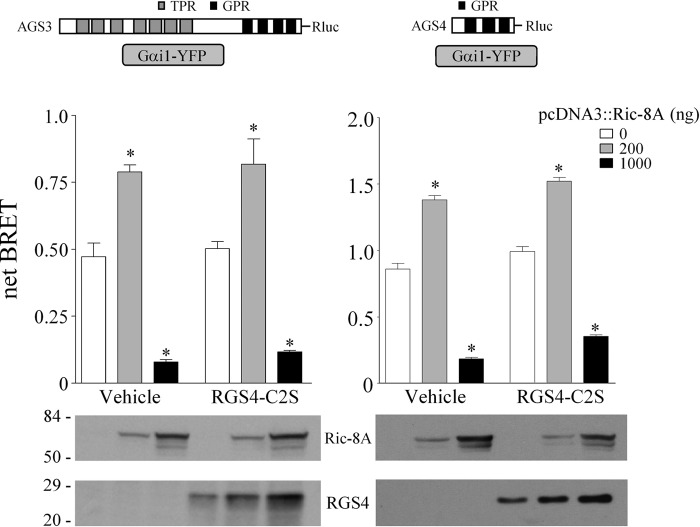
We also examined the effect of the GTPase accelerating protein RGS4 (RGS4-C2S) on Ric-8A regulation of the GPR-Gαi1 complex. The C2S mutation in RGS4 increases the stability of the protein (77). RGS4-C2S does not alter basal AGS3-Rluc-Gαi1-YFP and AGS4-Rluc-Gαi1-YFP BRET (Fig. 4) (42), and it had no effect on the Ric-8A-mediated regulation of the GPR-Gαi1 BRET signal (Fig. 4). In contrast, co-expression of RGS4-C2S actually counteracted the receptor-mediated regulation of AGS3-Rluc-Gαi1-YFP BRET, and this action required the GTPase-accelerating protein activity of RGS4 (42). Of interest, co-expression of Ric-8A also increased the levels of RGS4-C2S (Fig. 4, lower panels) suggesting the existence of additional signaling complexes or pathways involving Ric-8A, perhaps independent of the regulation of the GPR-Gαi1 module by Ric-8A.
FIGURE 4.
Effect of RGS4 on AGS3-Rluc-Gαi1-YFP and on AGS4-Rluc-Gαi1-YFP BRET. AGS3-Rluc (10 ng) (left panel) and AGS4-Rluc (2 ng) (right panel) were expressed in HEK cells with Gαi1-YFP (250 ng) in the presence and absence of Ric-8A, and BRET was measured as described under “Experimental Procedures.” RGS4-C2S (500 ng) was co-expressed as indicated. Data are presented as the mean ± S.E. from four experiments with triplicate determinations. *, p < 0.05 compared with their control. Lower panel, Ric-8A and RGS4 immunoblot. Each lane contains 50 μg of total protein, and the immunoblot is representative of two separate experiments.
Influence of Ric-8A on GPR-Gαi1 BRET and the Distribution of AGS3, AGS4, and Gαi1 Following Cell Fractionation
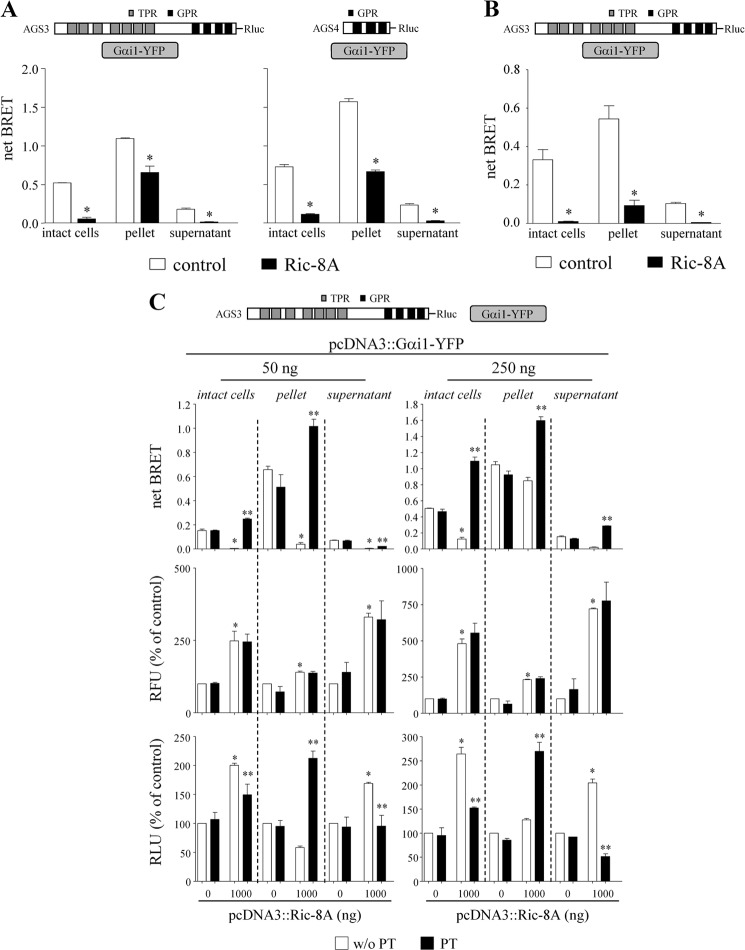
The data presented here clearly indicate that Ric-8A regulates the GPR-Gαi module in the intact cell. However, it is not known how these proteins position themselves in the cell and how signals may be transmitted to and sensed by Ric-8A and the GPR-Gα module. As one approach to this question, we asked if GPR-Gαi BRET is observed in both pellet and supernatant fractions and, if so, was the BRET signal differentially regulated by Ric-8A?
In this series of experiments, the fluorescent, luminescent, and BRET signals were obtained, in parallel, from intact cells and from pellet and supernatant subcellular fractions prepared following cell lysis. Cells were lysed in hypotonic buffer. BRET measurements were obtained from these two fractions following addition of substrate. This approach also allowed us to determine the relative distribution of Gαi1-YFP and AGS3-Rluc or AGS4-Rluc in the two subcellular fractions and how this distribution may be influenced by Ric-8A. AGS3-Rluc-Gαi1-YFP BRET and AGS4-Rluc-Gαi1-YFP BRET were observed in both the pellet and supernatant fractions, and this interaction was regulated by Ric-8A in a manner that mirrored the biphasic and concentration-dependent regulation of AGS3-Rluc-Gαi1-YFP BRET and AGS4-Rluc-Gαi1-YFP BRET observed in the intact cell (Fig. 5A).9 However, the magnitude of the BRET signal was much greater in the pellet fraction as compared with the supernatant fraction despite similar or even greater levels of AGS3-Rluc or AGS4-Rluc and Gαi1-YFP in the supernatant fractions (Table 2). The subcellular distribution of AGS3-Rluc-Gαi1-YFP BRET and the action of Ric-8A in HEK cells was similar to that observed in the neuronal catecholaminergic cell line (CAD) (Fig. 5B and Table 2). These data are consistent with the idea that the interaction of Gαi1-YFP with AGS3-Rluc or AGS4-Rluc stabilizes the proteins at the plasma membrane. Nevertheless, significant AGS3-Rluc-Gαi1-YFP BRET and AGS4-Rluc-Gαi1-YFP BRET were observed in the supernatant fraction.
FIGURE 5.
Ric-8A regulates AGS3-Rluc-Gαi1-YFP and AGS4-Rluc-Gαi1-YFP BRET in both pellet and supernatant fractions. A, AGS3-Rluc (10 ng) and AGS4-Rluc (2 ng) were expressed in HEK cells with Gαi1-YFP (250 ng), and BRET was measured in intact cells, pellet, and supernatant as described under “Experimental Procedures.” Ric-8A was expressed as indicated. Results are expressed as the mean ± S.E. of three independent experiments with triplicate determinations. *, p < 0.05 compared with their control. B, AGS3-Rluc (10 ng) was expressed in the neuronal catecholaminergic cell line (CAD) together with Gαi1-YFP (50 ng) and Ric-8A as indicated, and BRET was measured as described under “Experimental Procedures.” Data are presented as the mean ± S.E. from three experiments with triplicate determinations. *, p < 0.05 compared with their control as determined by Student's t test. The relative fluorescent units and relative luciferase units for sample points in A and B are presented in Table 2. C, AGS3-Rluc (10 ng) and Gαi1-YFP (50 ng) (left panel) or Gαi1-YFP (250 ng) (right panel) in the presence and absence of Ric-8A (1,000 ng) were expressed in HEK cells, and BRET was measured in intact cells, pellet, and supernatant fractions prepared from control cells or cells pretreated with PT as described under “Experimental Procedures.” RFU and RLU are presented as the percentage of control values obtained in cells expressing only AGS3-Rluc and Gαi1-YFP. Results are expressed as the mean ± S.E. of two independent experiments with triplicate determinations. *, p < 0.05 Ric-8A versus control. **, p < 0.05 PT versus non-PT treated control. Basal levels of RFU and RLU for control cells were as follows: 50 ng of pcDNA3::Gαi1-YFP: intact cell RFU = 89,412 ± 837, RLU = 524,526 ± 115,170; pellet RFU = 61,037 ± 9,021, RLU = 182,237 ± 82,483; supernatant RFU = 28,810 ± 7,388, RLU = 455,064 ± 255,751. 250 ng of pcDNA3::Gαi1-YFP: intact cell RFU = 141,573 ± 6,692, RLU = 338,131 ± 75,783; pellet RFU = 113,476 ± 11,240, RLU = 204,852 ± 76,241; supernatant RFU = 41,276 ± 9,572, RLU = 204,829 ± 110,133.
TABLE 2.
Influence of Ric-8A on the expression levels of AGS3-Rluc, AGS4-Rluc, and Gαi1-YFP in intact cells and lysed cell fractions
RFU and RLU generated for the data set presented in Fig. 5 were measured as described under “Experimental Procedures” and expressed as percent of control values obtained in the absence of pcDNA3::Ric-8A transfection. Fig. 5A (left panel) RFU control values for intact cells, pellet, and supernatant were 135,611 ± 8,324, 108,948 ± 7,453, and 41,606 ± 3,633, respectively. Fig. 5A (left panel) RLU control values for intact cells, pellet, and supernatant were 279,532 ± 55,954, 167,690 ± 60,590, and 122,392 ± 42,200, respectively. Fig. 5A (right panel) RFU control values for intact cells, pellet, and supernatant were 135,611 ± 8,324, 108,948 ± 7,453, and 41,606 ± 3,633, respectively. Fig. 5A (right panel) RLU control values for intact cells, pellet, and supernatant were 250,528 ± 8,659, 132,706 ± 24,573, and 148,439 ± 11,220, respectively. Fig. 5B RFU control values for intact cells, pellet, and supernatant were 23,045 ± 2,808, 29,560 ± 3,299, and 17,530 ± 816, respectively. Fig. 5B RLU control values for intact cells, pellet, and supernatant were 37,396 ± 1,360, 48,946 ± 8,693, 96,874 ± 13,848, respectively. Results are expressed as the mean ± S.E. of four independent experiments with triplicate determinations.
| Intact cell | Pellet | Supernatant | ||
|---|---|---|---|---|
| Fig. 5A (left panel) | RFU | 423 ± 27a | 194 ± 34a | 608 ± 97a |
| RLU | 263 ± 34a | 62 ± 10a | 186 ± 12a | |
| Fig. 5A (right panel) | RFU | 574 ± 28a | 232 ± 4a | 708 ± 25a |
| RLU | 203 ± 14a | 100 ± 6 | 153 ± 13a | |
| Fig. 5B | RFU | 225 ± 19a | 136 ± 28a | 321 ± 49a |
| RLU | 239 ± 48a | 106 ± 38 | 212 ± 8a | |
a p < 0.05 compared with control values was determined by Student's t test.
As the magnitude of the Ric-8A regulation of GPR-Gαi BRET depends upon the relative expression of the proteins, we expanded these studies to include a lower concentration of Gαi1 and to examine the effect of Ric-8A on the subcellular distribution of Gαi and AGS3 in the pellet and supernatant. As indicated earlier, the inhibitory action of Ric-8A on AGS3-Rluc-Gαi1-YFP BRET predominated at the lower Gα expression level (Fig. 5C). Ric-8A increased the levels of Gαi1-YFP in the intact cell at both levels of transfected Gα; this increase was distributed to both the pellet and supernatant fractions with a notable preference for the supernatant fraction (Fig. 5C and Tables 1 and 2). The effect of Ric-8A on AGS3-Rluc-Gαi1-YFP BRET and AGS4-Rluc-Gαi1-YFP BRET was mirrored by altered distribution of AGS3-Rluc and AGS4-Rluc in the pellet and supernatant fractions (Fig. 5, A and C, and Table 2). In circumstances where Ric-8A reduced the amount of AGS3-Rluc or AGS4-Rluc in the pellet fraction, there were corresponding increases in the amount of AGS3-Rluc and AGS4-Rluc in the supernatant fraction.
We then examined the effect of PT pretreatment on the altered distribution of the GPR proteins in the pellet and supernatant fractions. PT treatment did not alter the distribution of Gαi1-YFP in the presence or absence of Ric-8A. The subcellular re-distribution of AGS3-Rluc to the supernatant upon co-expression of Ric-8A was reversed by cell pretreatment with PT (Fig. 5C) resulting in a marked increase in the amount of both AGS3-Rluc and the magnitude of the AGS3-Rluc-Gαi1-YFP BRET in the pellet fraction at both levels of Gα expression (Fig. 5C).
Role of the Carboxyl Terminus of Ric-8A in the Regulation of Gα and the GPR-Gα Complex by Ric-8A
As a first approach to defining domains of Ric-8A required for the observed bioactivity, we examined the effect of carboxyl-terminal truncations on the Ric-8A-mediated increases in Gα and the Ric-8A-mediated regulation of the GPR-Gαi1 signaling cassette. Ric-8A contains multiple helical domains (78) and is predicted to contain 10 armadillo repeats (79). Ric-8A (Met1–Asn492) lacks a carboxyl-terminal helical domain, whereas Ric-8A (Met1–Asn453) lacks two predicted helical domains. Purified Ric-8A (Met1–Asn492) was a more robust GEF than full-length Ric-8A in promoting purified Gαi1-GDP release and GTPγS binding (78). Purified Ric-8A (Met1–Asn453) actually exhibited less GEF activity than full-length Ric-8A promoting GDP release but lacking any effect on GTPγS binding with purified Gαi1 (78). We first examined the effect of carboxyl-terminal truncations on the ability of Ric-8A to restore steady-state levels of Gαi and Gαq in Ric-8A−/− ES cells and the increased levels of Gαi1-YFP observed upon co-expression of Ric-8A in HEK cells. Expression of Ric-8A (Met1–Asn492), but not Ric-8A (Met1–Asn453), partially complemented the defect in Gα expression observed in Ric-8A−/− ES cells (Fig. 6A). Similarly, Ric-8A (Met1–Asn492) exhibited a reduced ability to increase Gαi1-YFP levels in HEK cells, and Ric-8A (Met1–Asn453) had no effect on Gαi1-YFP levels (Fig. 6B).10 These data indicate that the Ric-8A carboxyl terminus is an important domain with respect to its role in the regulation of steady-state levels of Gα in the cell. The region of Ric-8A between Asn453 and Asn493 appears critical for Ric-8A to increase Gαi expression levels. The inability of Ric-8A (Met1–Asn453) to complement the steady-state defect in Gα expression Ric-8A−/− ES cells may relate to its apparent inability to promote both GDP dissociation and binding of GTP (78). In the latter situation, the full cycle of nucleotide exchange would not be completed, which may be required for stabilization of Gα in the cell as suggested by Gabay et al. (66).
FIGURE 6.
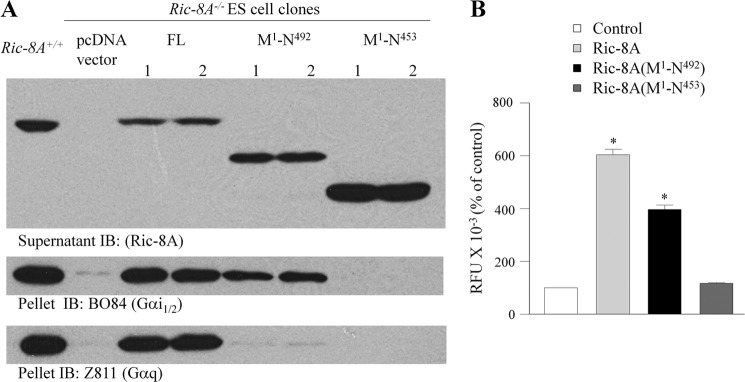
Effect of carboxyl-terminal truncated Ric-8A on Gα expression. A, complementation assay in Ric-8A−/− murine embryonic stem cells. Ric-8A−/− ES cell clones stably expressing full-length Ric-8A, Ric-8A (Met1–Asn492), Ric-8A (Met1–Asn453), or pcDNA3.1 Hygro were lysed and fractionated into pellet and supernatant as described elsewhere (66). Equal protein from the fractions was immunoblotted (IB) for Ric-8A, Gαi1/2 (B084), and Gαq/11 (Z811). B, AGS3-Rluc (10 ng), Gαi1-YFP (500 ng), and Ric-8A WT, Ric-8A (Met1–Asn453), and Ric-8A (Met1–Asn492) (1000 ng) in pcDNA3 were expressed in HEK cells, and relative fluorescence units (RFU) were measured as described under “Experimental Procedures.” Results are expressed as the mean ± S.E. of four independent experiments with triplicate determinations. *, p < 0.05 compared with control.
We then examined the role of the carboxy-terminal region on the Ric-8A-mediated regulation of the GPR-Gαi1 signaling cassette. Both Ric-8A (Met1–Asn453) and Ric-8A (Met1–Asn492) exhibited a reduced ability to inhibit AGS3-Rluc-Gαi1-YFP BRET (Fig. 7). As the magnitude of the Ric-8A regulation of GPR-Gαi1 BRET depends upon the relative expression of the proteins, we further examined the regulation of GPR-Gαi1 BRET by Ric-8A over a range of protein levels. Fig. 7A presents the data obtained with two different levels of Gα protein that illustrate this point, whereas the data obtained over a more complete range of Gα expression levels are presented as Fig. 7B. As observed for full-length Ric-8A, at low concentrations of Ric-8A (Met1–Asn492) the AGS3-Rluc-Gαi1-YFP BRET was inhibited, but at higher levels of Ric-8A (Met1–Asn492) the AGS3-Rluc-Gαi1-YFP BRET was augmented reflecting the increased expression of Gαi1-YFP (Fig. 7, B and C; Table 3). However, only the inhibitory component was observed upon expression of Ric-8A (Met1–Asn453) (Fig. 7, B and C). As observed for full-length Ric-8A, the inhibitory effect of the carboxyl-terminal truncated Ric-8A constructs was reduced or reversed by pertussis toxin treatment (Fig. 7C). These data suggest that even though Ric-8A (Met1–Asn453) did not increase Gαi1-YFP levels, it apparently retains some affinity for Gα consistent with its reported effects on purified Gαi1 (78).
FIGURE 7.
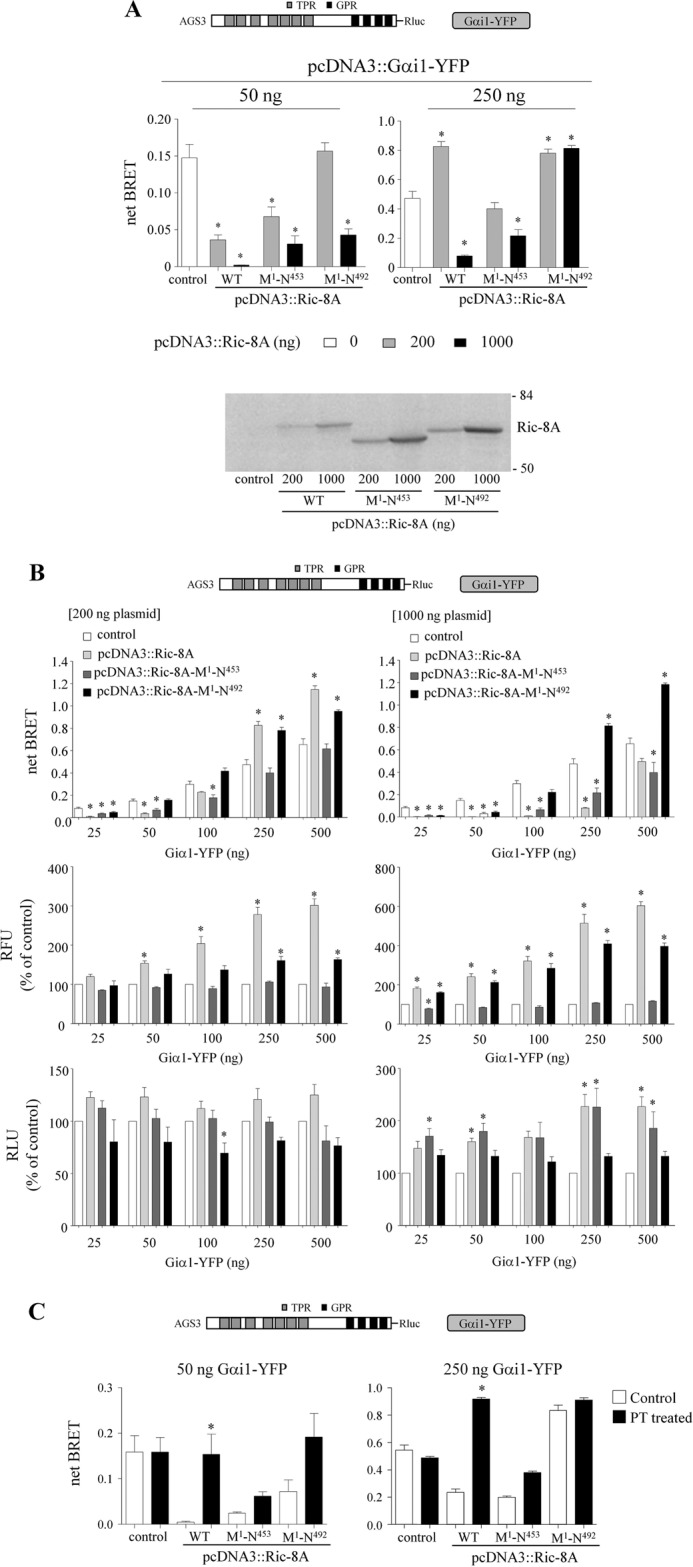
Effect of truncated Ric-8A on AGS3-Rluc-Gαi1-YFP BRET. A, BRET data presented in A were extracted from the larger, complete datasets in B. *, p < 0.05, compared with its control. Lower panel, Ric-8A immunoblot. Each lane contains 50 μg of total protein, and the immunoblot presented is representative of three separate experiments. The numbers to the right of the immunoblots correspond to the migration of prestained Bio-Rad protein standards. B, AGS3-Rluc (10 ng), increasing amounts of Gαi1-YFP (25–500 ng) and Ric-8A WT, Ric-8A (Met1–Asn453), and Ric-8A (Met1–Asn492) (as indicated in the figures) were expressed in HEK cells, and BRET signals were measured as described under “Experimental Procedures.” Middle panel, RFUs for each sample are presented as the percentage of control values (cells expressing only AGS3-Rluc and Gαi1-YFP). RFU values for control cells transfected with 25, 50, 100, 250, and 500 ng of pcDNA3::Gαi1-YFP 50,476 ± 6,117, 62,489 ± 6,986, 87,708 ± 11,705, 120,783 ± 16,267, and 155,140 ± 20,720, respectively. Lower panel, RLU are presented as the percentage of control values obtained in cells expressing only AGS3-Rluc and Gαi1-YFP. Relative luminescence unit values for control cells transfected with 25, 50, 100, 250, and 500 ng of pcDNA3::Gαi1-YFP were 462,452 ± 134,067, 456,049 ± 110,347, 400,833 ± 100,309, 263,295 ± 52,793, and 207,460 ± 45,087, respectively. Results are expressed as the mean ± S.E. of four independent experiments with triplicate determinations. *, p < 0.05 compared with their control. C, effect of PT pretreatment on the regulation of AGS3-Rluc-Gαi1-YFP BRET by carboxyl-terminal truncated Ric-8A. AGS3-Rluc (10 ng) and Gαi1-YFP (50 ng) (left panel) or Gαi1-YFP (250 ng) (right panel) were expressed in HEK cells and BRET signals, RFU and RLU values were measured as described under “Experimental Procedures.” The relative fluorescent units and relative luciferase units are presented in Table 3. Ric-8A and truncated Ric-8A mutants were expressed as indicated. In some experiments, cells were pretreated with pertussis toxin (100 ng/ml) for 16 h. Results are expressed as the mean ± S.E. of three independent experiments with triplicate determinations. *, p < 0.05 compared with their control.
TABLE 3.
Effect of pertussis toxin pretreatment on the expression of AGS3-Rluc and Gαi1-YFP in cells transfected with carboxyl-terminal truncated Ric-8A
RFU and RLU generated for the data set presented in Fig. 7C were measured as described under “Experimental Procedures” and expressed as % of control values observed in the absence of pcDNA3::Ric-8A transfection. Fig. 7C (left panel), RFU values for control and PT treatment were 57,640 ± 6,007 and 52,652 ± 4,983, respectively. Fig. 7C (left panel), RLU values for control and PT treatment were 613,083 ± 17,652 and 550,925 ± 19,720, respectively. Fig. 7C (right panel), RFU values for control and PT treatment were 105,582 ± 9,430 and 104,497 ± 10,406, respectively. Fig. 7C (right panel), RLU values for control and PT treatment were 357,334 ± 943 and 331,808 ± 13,914, respectively. Results are expressed as the mean ± S.E. of three independent experiments with triplicate determinations.
| Ric-8A-WT | Ric-8A-M1-N453 | Ric-8A-M1-N492 | ||
|---|---|---|---|---|
| Fig. 7C (left panel) | RFU | 206 ± 11 | 77 ± 1 | 150 ± 3 |
| RFU (PT) | 213 ± 11 | 89 ± 1 | 159 ± 10 | |
| RLU | 158 ± 11 | 145 ± 11 | 151 ± 5 | |
| RLU (PT) | 134 ± 2 | 140 ± 10 | 114 ± 4 | |
| Fig. 7C (right panel) | RFU | 528 ± 39 | 114 ± 7 | 313 ± 16 |
| RFU (PT) | 544 ± 23 | 129 ± 3 | 315 ± 9 | |
| RLU | 226 ± 25 | 154 ± 15 | 146 ± 3 | |
| RLU (PT) | 177 ± 8 | 160 ± 7 | 139 ± 8 | |
Mechanistic Considerations
The overall data presented are consistent with the forward cycle of nucleotide exchange and hydrolysis proposed by Thomas et al. (40), in which Ric-8A acts upon a GPR-Gαi1 complex to promote nucleotide exchange and dissociation of Gαi1 from the GPR protein (39). Subsequent hydrolysis of the bound GTP generates Gαi-GDP for re-association with the GPR protein or perhaps Gβγ. The Gαβγ heterotrimer is not a substrate for Ric-8A, but the reformed GPR-Gαi1 complex could again serve as a substrate for Ric-8A. The reduced AGS3-Rluc-Gαi1-YFP or AGS4-Rluc-Gαi1-YFP BRET signals in the presence of Ric-8A likely reflect such a cycle at some degree of equilibrium. PT treatment in the presence of Ric-8A would gradually result in disruption of the cycle and the accumulation of ADP-ribosylated Gαi1-YFP bound to AGS3-Rluc or AGS4-Rluc, which is then manifested as an increase in AGS3-Rluc-Gαi1-YFP BRET or AGS4-Rluc-Gαi1-YFP BRET as compared with the signal observed without PT pretreatment (Figs. 2 and 5C). The magnitude of this increase would be an indirect indicator of the cycling kinetics. At low concentrations of Gαi1, Ric-8A is capable of effectively driving the system to shift the equilibrium such that minimal Gαi1 is complexed with AGS3-Rluc or AGS4-Rluc. However, as Gαi1 increases, the equilibrium favors the formation of the AGS3-Rluc-Gαi1-YFP or AGS4-Rluc-Gαi1-YFP complex. It is difficult to completely eliminate the possibility that the observed changes in GPR-Gαi1-YFP BRET reflect competition between Ric-8A and AGS3 for binding to Gαi, independent of Ric-8A GEF activity. In contrast to Ric-8A-mediated reduction in AGS3-Rluc- or AGS4-Rluc-Gαi1-YFP BRET, AGS1 and GIV were without effect despite their clear ability to bind Gαi in vitro, which, together with the action of Ric-8A as a GEF in vitro, indicates that the regulation of the GPR-Gαi complex by Ric-8A in the cell is not likely a matter of competition for Gα binding.
The timing and mechanism of the interaction of GPR proteins with Gαi1 and the Ric-8A-mediated reduction of GPR-Gαi1 BRET are of interest. As recently reported, both AGS3-Rluc-Gαi1-YFP and AGS4-Rluc-Gαi1-YFP BRET were also reduced by activation of a cell surface receptor. In this situation, interaction of Gαi1 with AGS3 or AGS4 translocates the protein to the plasma membrane where it senses receptor activation leading to apparent reversible “release” of the GPR protein from the plasma membrane (42, 43). Different scenarios may be operative with respect to the Ric-8A-mediated regulation of the GPR-Gαi1 complex as reported here. One possibility is that AGS3 or AGS4 complex with Gαi1-GDP co-translationally or shortly thereafter, and the complex is then acted upon by Ric-8A in the cytosol resulting in GPR-Gα dissociation before the complex localizes at the plasma membrane. A second possibility is that Ric-8A acts upon the GPR-Gαi1-GDP complex after it is localized or stabilized at the plasma membrane. Either possibility would lead to stimulation of Gαi1 nucleotide exchange and dissociation of Gαi1 from the GPR motif. Such regulation by Ric-8A would result in reduced AGS3 or AGS4 protein at the plasma membrane because Gαi1 plays an important role in cortical positioning of AGS3 and AGS4 (42, 43, 70, 80). A third possibility is that Ric-8A forms a stable complex with Gαi1 in the cytosol as the protein is translated before a GPR protein can bind to Gαi1. Purified Ric-8A and Gαi1 can indeed exist as a stable complex in the absence of added nucleotide, and a recombinant Ric-8A-Gαq complex was observed in the soluble fraction during expression in insect cells (75). However, within the cell one would imagine that the guanine nucleotide exchange activity of Ric-8A and the cellular levels of guanine nucleotides would lead to actual dissociation of the Ric-8A-Gαi1 complex unless other regulatory mechanisms were in play.
Each of these scenarios are consistent with published results and the data presented here. In the third scenario involving the formation of a Ric-8A-Gαi1 stable complex, Ric-8A may act as a chaperone to stabilize nascent Gα (66), which results in the increased levels of Gαi-YFP observed in this study. Our data suggest that Ric-8A works upon G-proteins at two distinct points of their life cycle. Ric-8A acts as a molecular chaperone to promote proper Gα levels and also acts as a guanine nucleotide exchange factor for GPR-Gαi complexes in the context of cellular signaling functions. The molecular chaperone activity of Ric-8A would be PT-insensitive as a Ric-8A-Gαi1 complex is not expected to be an effective substrate for PT. The Gαi1 bound to its chaperone Ric-8A would be “delivered” to a binding partner (e.g. GPR protein or Gβγ).11 The GPR-Gαi complex, but not the Gβγ complex, would be a target for Ric-8A as a guanine nucleotide exchange factor. Once bound to a binding partner, Gαi would be a suitable substrate for ADP-ribosylation by PT, and thus PT treatment would block the action of Ric-8A as a GEF for the GPR-Gαi. Such a working hypothesis is consistent with the results presented here and the biochemical and functional properties of Ric-8A and GPR proteins as defined in the literature. The results presented here are indicative of a dynamic interaction between the GPR-Gαi1 complex and Ric-8A in the cell that influences subcellular localization of the three proteins and regulated complex formation.
Acknowledgments
We thank Heather Bainbridge, Sarah Barry, and Christine Marking for technical assistance. We thank Drs. Mikel Garcia-Marcos and Marilyn Farquhar (Department of Cell Biology, University of California, San Diego) and Dr. Takahashi (Nagoya University, Nagoya, Japan) for the full-length GIV construct, Dr. Mary Cismowski (Nationwide Children's Hospital, Columbus, OH) for pcDNA3.1/His::AGS1, Dr. Richard Neubig (University of Michigan, Ann Arbor, MI) for pcDNA3::RGS4-C2S, and Dr. Michel Bouvier (Department of Biochemistry, Institute for Research in Immunology and Cancer, Université de Montréal, Montréal, Québec, Canada) for the Rluc plasmids.
This work was supported, in whole or in part, by National Institutes of Health Grants DA025896 (to S. M. L.), GM086510 (to J. B. B.), and GM088242 (to G. G. T.).
S. S. Oner and S. M. Lanier, unpublished observations.
A similar effect of Ric-8A was observed with RGS14-Gαi1 BRET experiments as described in Vellano et al. (46).
E. M. Maher and J. B. Blumer, unpublished observations.
S. S. Oner, E. M. Maher, J. B. Blumer, and S. M. Lanier, unpublished observations.
The effects of receptor activation and Ric-8A on GPR-Gαi1 BRET appear additive, and only the pellet-associated GPR-Gα BRET was regulated by receptor activation (S. S. Oner, J. B. Blumer, and S. M. Lanier, unpublished data).
RNAi-mediated knockdown of endogenous Ric-8A in HEK-293 cells also reduced Gαi1-YFP expression (S. S. Oner, J. B. Blumer, and S. M. Lanier, unpublished observations.
It is also possible that there is a larger assembly of a complex involving Ric-8A, the GPR protein, and Gαi. Ric-8A-YFP and AGS3-Rluc exhibit a low level of BRET that is dependent upon co-expression of Gαi1 (S. S. Oner and S. M. Lanier, unpublished observations). Such a complex was also immunoprecipitated from D. melanogaster embryonic extracts (Ric-8) with the AGS3 and LGN D. melanogaster ortholog Pins (60, 64).
- AGS
- activators of G-protein signaling
- PT
- pertussis toxin
- GEF
- guanine nucleotide exchange factor
- GPR
- G-protein regulatory
- BRET
- bioluminescence resonance energy transfer
- RFU
- relative fluorescence unit
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate
- RLU
- relative luminescence unit.
REFERENCES
- 1. Cismowski M. J., Takesono A., Ma C., Lizano J. S., Xie X., Fuernkranz H., Lanier S. M., Duzic E. (1999) Genetic screens in yeast to identify mammalian nonreceptor modulators of G-protein signaling. Nat. Biotechnol. 17, 878–883 [DOI] [PubMed] [Google Scholar]
- 2. Takesono A., Cismowski M. J., Ribas C., Bernard M., Chung P., Hazard S., 3rd, Duzic E., Lanier S. M. (1999) Receptor-independent activators of heterotrimeric G-protein signaling pathways. J. Biol. Chem. 274, 33202–33205 [DOI] [PubMed] [Google Scholar]
- 3. Blumer J. B., Smrcka A. V., Lanier S. M. (2007) Mechanistic pathways and biological roles for receptor-independent activators of G-protein signaling. Pharmacol. Ther. 113, 488–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. De Vries L., Fischer T., Tronchère H., Brothers G. M., Strockbine B., Siderovski D. P., Farquhar M. G. (2000) Activator of G-protein signaling 3 is a guanine dissociation inhibitor for Gαi subunits. Proc. Natl. Acad. Sci. U.S.A. 97, 14364–14369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Du Q., Stukenberg P. T., Macara I. G. (2001) A mammalian Partner of inscuteable binds NuMA and regulates mitotic spindle organization. Nat. Cell Biol. 3, 1069–1075 [DOI] [PubMed] [Google Scholar]
- 6. Garcia-Marcos M., Ghosh P., Farquhar M. G. (2009) GIV is a nonreceptor GEF for Gαi with a unique motif that regulates Akt signaling. Proc. Natl. Acad. Sci. U.S.A. 106, 3178–3183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gotta M., Ahringer J. (2001) Distinct roles for Gα and Gβγ in regulating spindle position and orientation in Caenorhabditis elegans embryos. Nat. Cell Biol. 3, 297–300 [DOI] [PubMed] [Google Scholar]
- 8. Lee M. J., Dohlman H. G. (2008) Coactivation of G-protein signaling by cell-surface receptors and an intracellular exchange factor. Curr. Biol. 18, 211–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Parmentier M. L., Woods D., Greig S., Phan P. G., Radovic A., Bryant P., O'Kane C. J. (2000) Rapsynoid/partner of inscuteable controls asymmetric division of larval neuroblasts in Drosophila. J. Neurosci. 20, RC84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sato M., Blumer J. B., Simon V., Lanier S. M. (2006) Accessory proteins for G-proteins. Partners in signaling. Annu. Rev. Pharmacol. Toxicol. 46, 151–187 [DOI] [PubMed] [Google Scholar]
- 11. Schaefer M., Shevchenko A., Shevchenko A., Knoblich J. A. (2000) A protein complex containing Inscuteable and the Gα-binding protein Pins orients asymmetric cell divisions in Drosophila. Curr. Biol. 10, 353–362 [DOI] [PubMed] [Google Scholar]
- 12. Tall G. G., Krumins A. M., Gilman A. G. (2003) Mammalian Ric-8A (synembryn) is a heterotrimeric Gα protein guanine nucleotide exchange factor. J. Biol. Chem. 278, 8356–8362 [DOI] [PubMed] [Google Scholar]
- 13. Weiss T. S., Chamberlain C. E., Takeda T., Lin P., Hahn K. M., Farquhar M. G. (2001) Gαi3 binding to calnuc on Golgi membranes in living cells monitored by fluorescence resonance energy transfer of green fluorescent protein fusion proteins. Proc. Natl. Acad. Sci. U.S.A. 98, 14961–14966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Willard F. S., Kimple R. J., Siderovski D. P. (2004) Return of the GDI. The GoLoco motif in cell division. Annu. Rev. Biochem. 73, 925–951 [DOI] [PubMed] [Google Scholar]
- 15. Yu F., Morin X., Cai Y., Yang X., Chia W. (2000) Analysis of partner of inscuteable, a novel player of Drosophila asymmetric divisions, reveals two distinct steps in inscuteable apical localization. Cell 100, 399–409 [DOI] [PubMed] [Google Scholar]
- 16. Blumer J. B., Sadik Oner S., Lanier S. M. (2011) Group II activators of G-protein signalling and proteins containing a G-protein regulatory motif. Acta Physiol. 204, 202–218 [DOI] [PubMed] [Google Scholar]
- 17. Blumer J. B., Lord K., Saunders T. L., Pacchioni A., Black C., Lazartigues E., Varner K. J., Gettys T. W., Lanier S. M. (2008) Activator of G-protein signaling 3 null mice. I. Unexpected alterations in metabolic and cardiovascular function. Endocrinology 149, 3842–3849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bowers M. S., Hopf F. W., Chou J. K., Guillory A. M., Chang S. J., Janak P. H., Bonci A., Diamond I. (2008) Nucleus accumbens AGS3 expression drives ethanol seeking through Gβγ. Proc. Natl. Acad. Sci. U.S.A. 105, 12533–12538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bowers M. S., McFarland K., Lake R. W., Peterson Y. K., Lapish C. C., Gregory M. L., Lanier S. M., Kalivas P. W. (2004) Activator of G-protein signaling 3. A gatekeeper of cocaine sensitization and drug seeking. Neuron 42, 269–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Groves B., Gong Q., Xu Z., Huntsman C., Nguyen C., Li D., Ma D. (2007) A specific role of AGS3 in the surface expression of plasma membrane proteins. Proc. Natl. Acad. Sci. U.S.A. 104, 18103–18108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee S. E., Simons S. B., Heldt S. A., Zhao M., Schroeder J. P., Vellano C. P., Cowan D. P., Ramineni S., Yates C. K., Feng Y., Smith Y., Sweatt J. D., Weinshenker D., Ressler K. J., Dudek S. M., Hepler J. R. (2010) RGS14 is a natural suppressor of both synaptic plasticity in CA2 neurons and hippocampal-based learning and memory. Proc. Natl. Acad. Sci. U.S.A. 107, 16994–16998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nadella R., Blumer J. B., Jia G., Kwon M., Akbulut T., Qian F., Sedlic F., Wakatsuki T., Sweeney W. E., Jr., Wilson P. D., Lanier S. M., Park F. (2010) Activator of G-protein signaling 3 promotes epithelial cell proliferation in PKD. J. Am. Soc. Nephrol. 21, 1275–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pattingre S., De Vries L., Bauvy C., Chantret I., Cluzeaud F., Ogier-Denis E., Vandewalle A., Codogno P. (2003) The G-protein regulator AGS3 controls an early event during macroautophagy in human intestinal HT-29 cells. J. Biol. Chem. 278, 20995–21002 [DOI] [PubMed] [Google Scholar]
- 24. Regner K. R., Nozu K., Lanier S. M., Blumer J. B., Avner E. D., Sweeney W. E., Jr., Park F. (2011) Loss of activator of G-protein signaling 3 impairs renal tubular regeneration following acute kidney injury in rodents. FASEB J. 25, 1844–1855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sans N., Wang P. Y., Du Q., Petralia R. S., Wang Y. X., Nakka S., Blumer J. B., Macara I. G., Wenthold R. J. (2005) mPins modulates PSD-95 and SAP102 trafficking and influences NMDA receptor surface expression. Nat. Cell Biol. 7, 1079–1090 [DOI] [PubMed] [Google Scholar]
- 26. Walsh T., Shahin H., Elkan-Miller T., Lee M. K., Thornton A. M., Roeb W., Abu Rayyan A., Loulus S., Avraham K. B., King M. C., Kanaan M. (2010) Whole exome sequencing and homozygosity mapping identify mutation in the cell polarity protein GPSM2 as the cause of nonsyndromic hearing loss DFNB82. Am. J. Hum. Genet. 87, 90–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wiser O., Qian X., Ehlers M., Ja W. W., Roberts R. W., Reuveny E., Jan Y. N., Jan L. Y. (2006) Modulation of basal and receptor-induced GIRK potassium channel activity and neuronal excitability by the mammalian PINS homolog LGN. Neuron 50, 561–573 [DOI] [PubMed] [Google Scholar]
- 28. Yao L., McFarland K., Fan P., Jiang Z., Inoue Y., Diamond I. (2005) Activator of G-protein signaling 3 regulates opiate activation of protein kinase A signaling and relapse of heroin-seeking behavior. Proc. Natl. Acad. Sci. U.S.A. 102, 8746–8751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yao L., McFarland K., Fan P., Jiang Z., Ueda T., Diamond I. (2006) Adenosine A2a blockade prevents synergy between μ-opiate and cannabinoid CB1 receptors and eliminates heroin-seeking behavior in addicted rats. Proc. Natl. Acad. Sci. U.S.A. 103, 7877–7882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ponting C. P. (1999) Raf-like Ras/Rap-binding domains in RGS12- and still-life-like signalling proteins. J. Mol. Med. 77, 695–698 [DOI] [PubMed] [Google Scholar]
- 31. Siderovski D. P., Diversé-Pierluissi Ma, De Vries L. (1999) The GoLoco motif. A Gαi/o binding motif and potential guanine-nucleotide exchange factor. Trends Biochem. Sci. 24, 340–341 [DOI] [PubMed] [Google Scholar]
- 32. An N., Blumer J. B., Bernard M. L., Lanier S. M. (2008) The PDZ and band 4.1 containing protein Frmpd1 regulates the subcellular location of activator of G-protein signaling 3 and its interaction with G-proteins. J. Biol. Chem. 283, 24718–24728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Blumer J. B., Bernard M. L., Peterson Y. K., Nezu J., Chung P., Dunican D. J., Knoblich J. A., Lanier S. M. (2003) Interaction of activator of G-protein signaling 3 (AGS3) with LKB1, a serine/threonine kinase involved in cell polarity and cell cycle progression. Phosphorylation of the G-protein regulatory (GPR) motif as a regulatory mechanism for the interaction of GPR motifs with Giα. J. Biol. Chem. 278, 23217–23220 [DOI] [PubMed] [Google Scholar]
- 34. Bowman S. K., Neumüller R. A., Novatchkova M., Du Q., Knoblich J. A. (2006) The Drosophila NuMA Homolog Mud regulates spindle orientation in asymmetric cell division. Dev. Cell 10, 731–742 [DOI] [PubMed] [Google Scholar]
- 35. Du Q., Macara I. G. (2004) Mammalian Pins is a conformational switch that links NuMA to heterotrimeric G-proteins. Cell 119, 503–516 [DOI] [PubMed] [Google Scholar]
- 36. Garcia-Marcos M., Ear J., Farquhar M. G., Ghosh P. (2011) A GDI (AGS3) and a GEF (GIV) regulate autophagy by balancing G-protein activity and growth factor signals. Mol. Biol. Cell 22, 673–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Johnston C. A., Hirono K., Prehoda K. E., Doe C. Q. (2009) Identification of an Aurora-A/PinsLINKER/Dlg spindle orientation pathway using induced cell polarity in S2 cells. Cell 138, 1150–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kopein D., Katanaev V. L. (2009) Drosophila GoLoco-protein Pins is a target of Gαo-mediated G-protein-coupled receptor signaling. Mol. Biol. Cell 20, 3865–3877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tall G. G., Gilman A. G. (2005) Resistance to inhibitors of cholinesterase 8A catalyzes release of Gαi-GTP and nuclear mitotic apparatus protein (NuMA) from NuMA/LGN/Gαi-GDP complexes. Proc. Natl. Acad. Sci. U.S.A. 102, 16584–16589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Thomas C. J., Tall G. G., Adhikari A., Sprang S. R. (2008) Ric-8A catalyzes guanine nucleotide exchange on Gαi1 bound to the GPR/GoLoco exchange inhibitor AGS3. J. Biol. Chem. 283, 23150–23160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vellano C. P., Shu F. J., Ramineni S., Yates C. K., Tall G. G., Hepler J. R. (2011) Activation of the regulator of G-protein signaling 14-Gαi1-GDP signaling complex is regulated by resistance to inhibitors of cholinesterase-8A. Biochemistry 50, 752–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Oner S. S., An N., Vural A., Breton B., Bouvier M., Blumer J. B., Lanier S. M. (2010) Regulation of the AGS3·Gαi signaling complex by a seven-transmembrane span receptor. J. Biol. Chem. 285, 33949–33958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Oner S. S., Maher E. M., Breton B., Bouvier M., Blumer J. B. (2010) Receptor-regulated interaction of activator of G-protein signaling-4 and Gαi. J. Biol. Chem. 285, 20588–20594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cismowski M. J., Ma C., Ribas C., Xie X., Spruyt M., Lizano J. S., Lanier S. M., Duzic E. (2000) Activation of heterotrimeric G-protein signaling by a Ras-related protein. Implications for signal integration. J. Biol. Chem. 275, 23421–23424 [DOI] [PubMed] [Google Scholar]
- 45. Hiskens R., Vatish M., Hill C., Davey J., Ladds G. (2005) Specific in vivo binding of activator of G-protein signalling 1 to the Gβ1 subunit. Biochem. Biophys. Res. Commun. 337, 1038–1046 [DOI] [PubMed] [Google Scholar]
- 46. Vellano C. P., Maher E. M., Hepler J. R., Blumer J. B. (2011) G-protein-coupled receptors and resistance to inhibitors of cholinesterase-8A (Ric-8A) both regulate the regulator of G-protein signaling 14 RGS14·Gαi1 complex in live cells. J. Biol. Chem. 286, 38659–38669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Graham T. E., Prossnitz E. R., Dorin R. I. (2002) Dexras1/AGS-1 inhibits signal transduction from the Gi-coupled formyl peptide receptor to Erk-1/2 MAP kinases. J. Biol. Chem. 277, 10876–10882 [DOI] [PubMed] [Google Scholar]
- 48. Nguyen C. H., Watts V. J. (2005) Dexras1 blocks receptor-mediated heterologous sensitization of adenylyl cyclase 1. Biochem. Biophys. Res. Commun. 332, 913–920 [DOI] [PubMed] [Google Scholar]
- 49. Vaidyanathan G., Cismowski M. J., Wang G., Vincent T. S., Brown K. D., Lanier S. M. (2004) The Ras-related protein AGS1/RASD1 suppresses cell growth. Oncogene 23, 5858–5863 [DOI] [PubMed] [Google Scholar]
- 50. Baniwal S. K., Khalid O., Gabet Y., Shah R. R., Purcell D. J., Mav D., Kohn-Gabet A. E., Shi Y., Coetzee G. A., Frenkel B. (2010) Runx2 transcriptome of prostate cancer cells. Insights into invasiveness and bone metastasis. Mol. Cancer 9, 258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dalgin G. S., Holloway D. T., Liou L. S., DeLisi C. (2007) Identification and characterization of renal cell carcinoma gene markers. Cancer Inform. 3, 65–92 [PMC free article] [PubMed] [Google Scholar]
- 52. de Souza Rocha Simonini P., Breiling A., Gupta N., Malekpour M., Youns M., Omranipour R., Malekpour F., Volinia S., Croce C. M., Najmabadi H., Diederichs S., Sahin O., Mayer D., Lyko F., Hoheisel J. D., Riazalhosseini Y. (2010) Epigenetically deregulated microRNA-375 is involved in a positive feedback loop with estrogen receptor α in breast cancer cells. Cancer Res. 70, 9175–9184 [DOI] [PubMed] [Google Scholar]
- 53. Nojima M., Maruyama R., Yasui H., Suzuki H., Maruyama Y., Tarasawa I., Sasaki Y., Asaoku H., Sakai H., Hayashi T., Mori M., Imai K., Tokino T., Ishida T., Toyota M., Shinomura Y. (2009) Genomic screening for genes silenced by DNA methylation revealed an association between RASD1 inactivation and dexamethasone resistance in multiple myeloma. Clin. Cancer Res. 15, 4356–4364 [DOI] [PubMed] [Google Scholar]
- 54. Shaw E. J., Haylock B., Husband D., du Plessis D., Sibson D. R., Warnke P. C., Walker C. (2011) Gene expression in oligodendroglial tumors. Cell Oncol. 34, 355–367 [DOI] [PubMed] [Google Scholar]
- 55. Ghosh P., Beas A. O., Bornheimer S. J., Garcia-Marcos M., Forry E. P., Johannson C., Ear J., Jung B. H., Cabrera B., Carethers J. M., Farquhar M. G. (2010) A Gαi-GIV molecular complex binds epidermal growth factor receptor and determines whether cells migrate or proliferate. Mol. Biol. Cell 21, 2338–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Garcia-Marcos M., Jung B. H., Ear J., Cabrera B., Carethers J. M., Ghosh P. (2011) Expression of GIV/Girdin, a metastasis-related protein, predicts patient survival in colon cancer. FASEB J. 25, 590–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Afshar K., Willard F. S., Colombo K., Johnston C. A., McCudden C. R., Siderovski D. P., Gönczy P. (2004) RIC-8 is required for GPR-1/2-dependent Gα function during asymmetric division of C. elegans embryos. Cell 119, 219–230 [DOI] [PubMed] [Google Scholar]
- 58. Couwenbergs C., Spilker A. C., Gotta M. (2004) Control of embryonic spindle positioning and Gα activity by C. elegans RIC-8. Curr. Biol. 14, 1871–1876 [DOI] [PubMed] [Google Scholar]
- 59. David N. B., Martin C. A., Segalen M., Rosenfeld F., Schweisguth F., Bellaïche Y. (2005) Drosophila Ric-8 regulates Gαi cortical localization to promote Gαi-dependent planar orientation of the mitotic spindle during asymmetric cell division. Nat. Cell Biol. 7, 1083–1090 [DOI] [PubMed] [Google Scholar]
- 60. Hampoelz B., Hoeller O., Bowman S. K., Dunican D., Knoblich J. A. (2005) Drosophila Ric-8 is essential for plasma-membrane localization of heterotrimeric G-proteins. Nat. Cell Biol. 7, 1099–1105 [DOI] [PubMed] [Google Scholar]
- 61. Hess H. A., Röper J. C., Grill S. W., Koelle M. R. (2004) RGS-7 completes a receptor-independent heterotrimeric G-protein cycle to asymmetrically regulate mitotic spindle positioning in C. elegans. Cell 119, 209–218 [DOI] [PubMed] [Google Scholar]
- 62. Miller K. G., Emerson M. D., McManus J. R., Rand J. B. (2000) RIC-8 (Synembryn). A novel conserved protein that is required for Gqα signaling in the C. elegans nervous system. Neuron 27, 289–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Miller K. G., Rand J. B. (2000) A role for RIC-8 (Synembryn) and GOA-1 (Goα) in regulating a subset of centrosome movements during early embryogenesis in Caenorhabditis elegans. Genetics 156, 1649–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wang H., Ng K. H., Qian H., Siderovski D. P., Chia W., Yu F. (2005) Ric-8 controls Drosophila neural progenitor asymmetric division by regulating heterotrimeric G-proteins. Nat. Cell Biol. 7, 1091–1098 [DOI] [PubMed] [Google Scholar]
- 65. Woodard G. E., Huang N. N., Cho H., Miki T., Tall G. G., Kehrl J. H. (2010) Ric-8A and Giα recruit LGN, NuMA, and dynein to the cell cortex to help orient the mitotic spindle. Mol. Cell. Biol. 30, 3519–3530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gabay M., Pinter M. E., Wright F. A., Chan P., Murphy A. J., Valenzuela D. M., Yancopoulos G. D., Tall G. G. (2011) Ric-8 proteins are molecular chaperones that direct nascent G-protein α subunit membrane association. Sci. Signal. 4, ra79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Audet N., Galés C., Archer-Lahlou E., Vallières M., Schiller P. W., Bouvier M., Pineyro G. (2008) Bioluminescence resonance energy transfer assays reveal ligand-specific conformational changes within preformed signaling complexes containing δ-opioid receptors and heterotrimeric G-proteins. J. Biol. Chem. 283, 15078–15088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Galés C., Rebois R. V., Hogue M., Trieu P., Breit A., Hébert T. E., Bouvier M. (2005) Real-time monitoring of receptor and G-protein interactions in living cells. Nat. Methods 2, 177–184 [DOI] [PubMed] [Google Scholar]
- 69. Galés C., Van Durm J. J., Schaak S., Pontier S., Percherancier Y., Audet M., Paris H., Bouvier M. (2006) Probing the activation-promoted structural rearrangements in preassembled receptor-G-protein complexes. Nat. Struct. Mol. Biol. 13, 778–786 [DOI] [PubMed] [Google Scholar]
- 70. Willard F. S., Zheng Z., Guo J., Digby G. J., Kimple A. J., Conley J. M., Johnston C. A., Bosch D., Willard M. D., Watts V. J., Lambert N. A., Ikeda S. R., Du Q., Siderovski D. P. (2008) A point mutation to Gαi selectively blocks GoLoco motif binding. Direct evidence for Gα·GoLoco complexes in mitotic spindle dynamics. J. Biol. Chem. 283, 36698–36710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Chauhan S., Jelen F., Sharina I., Martin E. (2012) The G-protein regulator LGN modulates the activity of the NO receptor soluble guanylate cyclase. Biochem. J. 446, 445–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Groves B., Abrahamsen H., Clingan H., Frantz M., Mavor L., Bailey J., Ma D. (2010) An inhibitory role of the G-protein regulator AGS3 in mTOR-dependent macroautophagy. PLoS ONE 5, e8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hofler C., Koelle M. R. (2011) AGS-3 alters Caenorhabditis elegans behavior after food deprivation via RIC-8 activation of the neural G-protein Gαo. J. Neurosci. 31, 11553–11562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Doherty D., Chudley A. E., Coghlan G., Ishak G. E., Innes A. M., Lemire E. G., Rogers R. C., Mhanni A. A., Phelps I. G., Jones S. J., Zhan S. H., Fejes A. P., Shahin H., Kanaan M., Akay H., Tekin M., FORGE Canada Consortium, Triggs-Raine B., Zelinski T. (2012) GPSM2 mutations cause the brain malformations and hearing loss in Chudley-McCullough syndrome. Am. J. Hum. Genet. 90, 1088–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Chan P., Gabay M., Wright F. A., Kan W., Oner S. S., Lanier S. M., Smrcka A. V., Blumer J. B., Tall G. G. (2011) Purification of heterotrimeric G-protein α subunits by GST-Ric-8 association. Primary characterization of purified Gαolf. J. Biol. Chem. 286, 2625–2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Casey P. J., Graziano M. P., Gilman A. G. (1989) G-protein βγ subunits from bovine brain and retina: equivalent catalytic support of ADP-ribosylation of α subunits by pertussis toxin but differential interactions with Gsα. Biochemistry 28, 611–616 [DOI] [PubMed] [Google Scholar]
- 77. Bodenstein J., Sunahara R. K., Neubig R. R. (2007) N-terminal residues control proteasomal degradation of RGS2, RGS4, and RGS5 in human embryonic kidney 293 cells. Mol. Pharmacol. 71, 1040–1050 [DOI] [PubMed] [Google Scholar]
- 78. Thomas C. J., Briknarová K., Hilmer J. K., Movahed N., Bothner B., Sumida J. P., Tall G. G., Sprang S. R. (2011) The nucleotide exchange factor Ric-8A is a chaperone for the conformationally dynamic nucleotide-free state of Gαi1. PLoS ONE 6, e23197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Figueroa M., Hinrichs M. V., Bunster M., Babbitt P., Martinez-Oyanedel J., Olate J. (2009) Biophysical studies support a predicted superhelical structure with armadillo repeats for Ric-8. Protein Sci. 18, 1139–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Vural A., Oner S., An N., Simon V., Ma D., Blumer J. B., Lanier S. M. (2010) Distribution of activator of G-protein signaling 3 within the aggresomal pathway. Role of specific residues in the tetratricopeptide repeat domain and differential regulation by the AGS3 binding partners Gi(α) and mammalian inscuteable. Mol. Cell. Biol. 30, 1528–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]