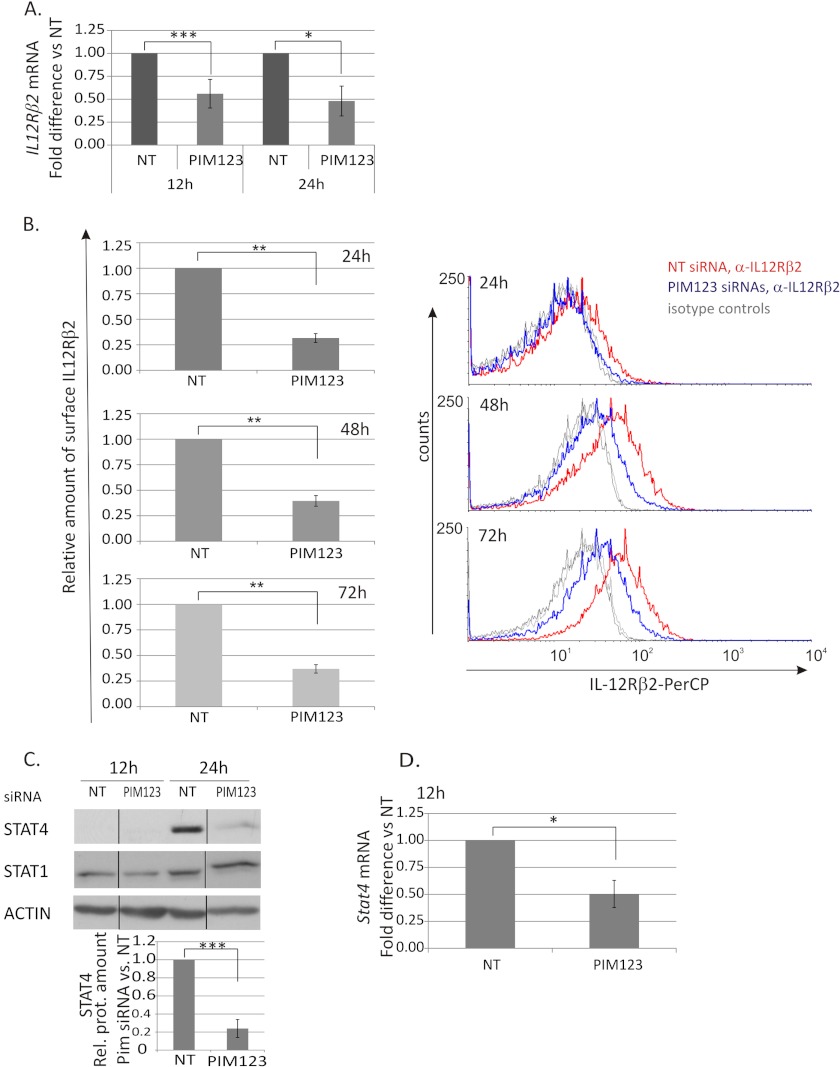
FIGURE 4.
PIM kinases regulate the IL-12Rβ2/STAT4 pathway in Th1-polarized cells. CD4+ cells were nucleofected and cultured as in Fig. 2. A, samples were harvested at different time points during the culture for TaqMan RT-PCR analysis. These graphs represent the average -fold differences ±S.E. (error bars) of IL-12Rβ2 mRNA levels in PIM siRNA-treated Th1 cells compared with NT siRNA-treated Th1 cells. Data were calculated from five of six independent experiments. B, expression of the cell surface IL-12Rβ2 protein was analyzed by immunofluorescent staining and flow cytometry at the indicated time points. The bars represent the average -fold changes ±S.E. (error bars) of the geometric mean intensities of IL-12Rβ2 in PIM siRNA samples compared with control samples. The data were calculated from six independent experiments. The histograms show representative data from these experiments (red line, NT siRNA Th1; blue line, PIM123 siRNA Th1; gray lines, isotype controls). Samples were harvested for Western blotting (C) and TaqMan RT-PCR (D) analyses at the indicated time points. The bars represent the mean values ±S.E. (error bars) of the relative levels of STAT4 between control and PIM siRNA samples obtained by quantifying and normalizing against the levels of β-actin. The value of control samples was set as 1. The data were calculated from four independent experiments. The Western blot of STAT1 shows representative data from four independent experiments. Vertical lines represent repositioned gel lanes that are from the same blot and the same exposure. A–D, statistical significances were calculated using the two-tailed paired t test: *, p < 0.05; **, p < 0.02; ***, p > 0.01 (A, C, and D); *, p < 0.005; **, p < 0.0005 (B). NT indicates control siRNA. Rel. prot., relative protein; PerCP, peridinin-chlorophyll protein complex.