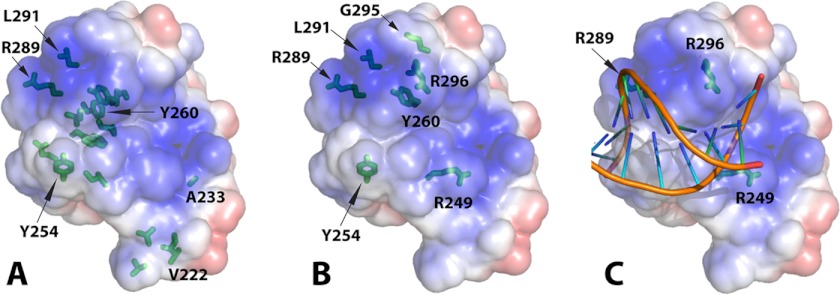
FIGURE 6.
VRN1(208–341)-DNA interaction. A, VRN1(208–341) in the same orientation as Fig. 3A and Fig. 5A displaying its electrostatic surface and marked with the residues most sensitive to NMR. The side chains of all residues in intermediate exchange at 0.2 m eq are shown in stick format, and those of the most sensitive residues in intermediate exchange at 0.1 m eq of DNA are also labeled textually. The figure with a slight rotation in supplemental Fig. S3 is better to show the “sidedness” of the DNA interaction. B, the side chains of VRN1(208–341) residues mutated preceding DNA binding studies are shown in stick format. C, the structure of VRN1(208–341) was superimposed onto the structure of EcoRII from the crystal structure of the EcoRII-DNA complex (Protein Data Bank code 3HQF) (30) using SSM in Coot. The structure of EcoRII was then removed, leaving this VRN1(208–341)-DNA overlay. No further manipulation was performed. PyMOL was used to produce the electrostatic surface of VRN1(208–341). The positions of three Arg residues (R249, R289, and R296) that, when mutated to Glu, abolished DNA binding are shown.