Abstract
The glial cells missing (gcm) regulatory gene of the sea urchin Strongylocentrotus purpuratus is first expressed in veg2 daughter cells as the genomic target of late cleavage stage Delta-Notch signaling from the skeletogenic mesoderm precursors. Gcm is required in veg2 progeny during late cleavages for the early phase of pigment cell precursor specification. Here we report on a later acting cis-regulatory module that assumes control of gcm expression by the early mesenchyme blastula stage and maintains it through pigment cell differentiation and dispersal. Cis-perturbation analyses reveal that the two critical elements within this late module are consensus matches to Gcm and Six1 binding sites. Significantly, six1 mRNA localizes to gcm+ cells from the mesenchyme blastula stage onwards. Trans-perturbations with anti-sense morpholinos reveal a co-dependency between six1 and gcm. Six1 mRNA levels fall sharply after Gcm is depleted, while depleting Six1 leads to significant reductions in output of endogenous gcm or modular-reporters. These results support the conclusion gcm and six1 comprise a positive intergenic feedback loop in the mesodermal GRN. This often employed cross regulatory GRN feature here ensures self-sustaining gcm output in a cohort of fully specified pigment cell precursors at a relatively early developmental stage.
Keywords: echinoid, pigment cell, mesoderm, cis-regulation, autoregulatory, intergenic feedback loop
Introduction
Echinoids have evolved mechanisms for early specification of mesodermal lineages that directly produce the skeletal spicules and pigment cells of the pluteus larva. The skeletogenic mesoderm is autonomously specified in the course of early vegetal unequal cleavages that yield the rigidly determined large micromere lineage. Following a stereotyped developmental program, the skeletogenic mesoderm precursors ingress as primary mesenchyme cells at the blastula stage, then form a syncytial array and begin secreting biomineralized larval skeletal spicules in late-gastrula stage embryos (Oliveri et al., 2008). By contrast, the initial specification of pigment cell precursors relies on non-autonomous cell interactions between large micromere and veg2 lineages. Initially, a non-skeletogenic mesoderm domain is specified during late cleavage stages in veg2 lineage cells that respond to Delta-ligand from the large micromeres (Sherwood and McClay, 1999; Sweet et al., 2002). During gastrulation the fully specified pigment cell precursors ingress and disperse to the aboral ectoderm (Gibson and Burke, 1985, 1987), although the precise timing varies considerably between echinoid species (Takata and Kominami, 2004). Differentiated pigment cells with echinochrome containing granules and ornate morphology are present soon after the completion of gastrulation. Thus, two fully differentiated mesodermal cell types are deployed at a relatively early stage in indirect developing echinoid embryos.
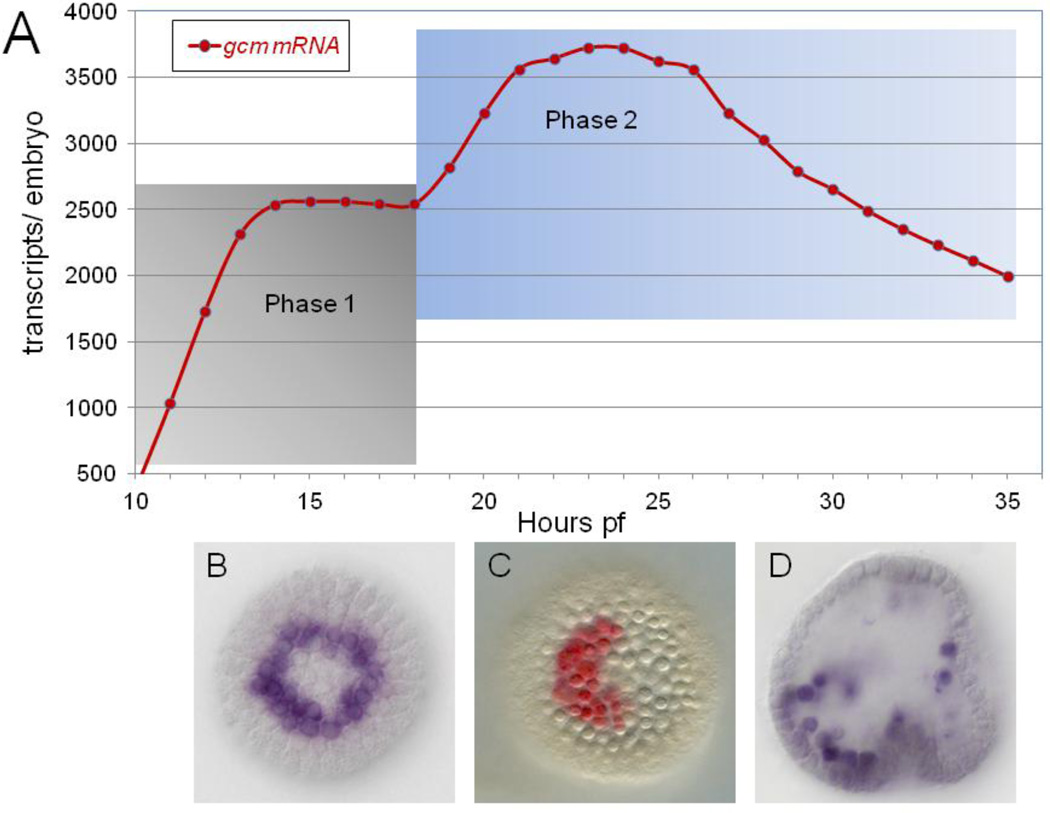
The non-skeletogenic mesoderm is first demarcated in embryos of the purple sea urchin Strongylocentrotus purpuratus (Sp) by expression of glial cell missing (gcm) (Ransick et al., 2002). Gcm is a direct target gene of Delta-Notch (DN) signaling that is essential for pigment cell specification (Ransick and Davidson, 2006). Gcm transcript levels rise from the onset of expression at the 7th cleavage stage until 9th cleavage stage (Materna et al., 2010). Here a distinct expression plateau is reached that persists into the late hatched blastula stage (Figs. 1A, S1). Throughout this initial phase of expression, a ring of veg2-derived gcm expressing cells (Fig. 1B) is maintained only through direct contact with Delta-expressing skeletogenic mesoderm (Ransick and Davidson, 2006; Croce and McClay, 2010).
Fig. 1.
Gcm expression profile A) high density mRNA time course. A portion of gcm expression profile from 7th cleavage onset near 10hpf through early gastrulation at 36hpf, distinguishing an early phase reliant on DN signaling (Phase 1, gray shading) from a second phase that extends from near PMC ingression through gastrulation (Phase 2, blue shading); data from Materna et al., 2010. An extended time course of gcm expression (through 48hpf) is shown in Fig. S1. B–D) gcm mRNA localized by whole mount in situ hybridization, showing a symmetric ring at 15hpf (B), aboral patch at 24hpf (C) and dispersing pigment cell precursors at 34hpf (D).
Yet, gcm expression clearly persists even after Delta levels drop in pre-ingression skeletogenic mesoderm (Oliveri et al., 2002; Sweet et al., 2002). Therefore this gene must be acquiring new regulatory inputs by the early mesenchyme blastula stage. Reflecting the new regulatory environment, gcm levels rise by fifty percent to peak levels in the mid-mesenchyme blastula. Also, the gcm+ cells now form a contiguous patch of ~30 cells centered on the aboral side of the vegetal plate (Fig. 1C). Gcm is necessary for maintenance of this cohort of pigment cell precursors within the vegetal plate throughout the ensuing period of complex specification processes that pattern other mesodermal cell types, the endoderm and the second axis of the embryo (Duboc et al, 2010; Peter 2011). Expression is also maintained during ingression and dispersal of fully specified pigment cell precursors (Fig. 1D), as Gcm is an important driver of the pigment differentiation gene battery (Calestani and Rogers, 2010).
Clearly, gcm expression at later stages (phase two in Fig. 1A) is governed by different cis-regulatory inputs than those driving the gene in cleavage stage embryos. Here we report analysis of a newly identified cis-regulatory module of gcm that becomes active in the early mesenchyme blastula and continues to drive expression through pigment cell differentiation. We find the cis-regulatory device encoded by the late module is a stabilizing intergenic loop (Davidson, 2009). The self-sustaining expression of gcm that the late module promotes effectively locks down the pigment cell fate.
Materials and methods
Reporter constructs and microinjections
Reporter construct microinjections were carried according to well-established protocols (McMahon et al., 1985); Arnone et al., 2004), using PCR products generated with insert specific primers from pGEM-T subclones of confirmed sequence. Microinjection solutions were prepared just prior to use and consisted of 10–25ng of construct DNA and 200ng HindIII digested genomic carrier DNA in 10 µl of 125 mM KCl. This formulation delivered 500–750 copies of the construct with a 7x molar excess of carrier DNA per two picoliter injections. The fluorescent protein coding sequences of Gfp and Rfp were derived from Green Lantern (Gibco) and mRfp1 (Shaner et al., 2004), respectively. Nuclearized Rfp (nRfp) was achieved with a 5′ in-frame insertion of the histone 2B fragment ahead of the mRfp1 start codon (p13-pCS-H2B-mRfp1 was a gift from Scott Fraser, Caltech). Recombinant BACs (Warming et al., 2005) included gcm::gfp [BAC 30-O18] with an exon 3 insertion of the gfp/SV40 cassette and six1::gfp [BAC 3058-B7] with insertion at the ATG site of exon 1.
Gcm reporters used here had two configurations relative to the region proximal to transcription start site. The original configuration, as previously described by Ransick & Davidson (2006), contained a longer proximal (P) module, that extended from bases −239 to +217 relative to the gcm transcription start, and was fused to an exogenous basal promoter fragment from EpGFPII (Arnone et al, 2004). In the alternative configuration, the fluorescent protein sequence was fused directly to a shortened P module containing bases −36 to +217 relative to gcm transcription start. The latter constitutes the minimal basal promoter gcm reporter and when used is indicated by a “P36” label.
Real-time PCR assays
Extraction of mRNA, processing, cDNA synthesis and real-time PCR assays (QPCR) were carried out on batches of 75–300 similarly treated embryos, according to our established protocols (Ransick, 2004; Oliveri and Davidson, 2004; Revilla-i-Domingo et al., 2004). In measurements of mRNA expression from cDNA samples, QPCR values for all gene specific primers were normalized to Ubiquitin values from that sample. Expression levels were converted to molecules per embryo, using as reference 45,000 copies of the ubq coding sequence per embryo after 20 hours post-fertilization (hpf) (Materna et al., 2010). To estimate the number of injected copies incorporated, QPCR values for gfp or rfp from genomic DNA samples were normalized to nodal values, as a single copy gene present at two copies per cell. A measure of output by an injected Gfp reporter is thus easily calculated in terms of mRNAs per incorporated copy, and this derivation of expression levels is used throughout this report.
Whole mount in situ hybridization
Whole mount in situ hybridization (WMISH) was performed as described by Ransick, 2004 with the following modifications: Fixation Buffer (FB) (after (Minokawa et al., 2004) consisted of 32.5% filtered sea water; 162.5 mM NaCl and 32.5 mM MOPS Buffer, pH 7.0. Fixation was carried out on ice, for 30min in FB plus 0.65% glutaraldehyde, followed by FB plus 1.25% glutaraldehyde for at least 3hrs (usually overnight). Hybridization buffer (HB) consisted of 50% deionized formamide, 5x SSC, 2x Denhardt’s, 5 mM EDTA, 20 mM Tris buffer, pH 7.5, 0.01% Tween-20, 0.5 mg/ml yeast tRNA and 50 µg/ml heparin. Hybridization was carried out overnight at 57°C (±2) with flourescein (Fl), digoxygenin (Dig) (Roche) or dinitrophenol (DNP) (Mirrus) modified antisense RNA probes at final concentrations of 0.3–1.0 ng/µl. The antisense probes for gfp (720nt) and six1 (1010nt) covered the entire coding sequences, while the probes for gcm (1000nt) and nfkb (1025nt) matched the 5’ ends and c-rel (840nt) the 3’ end of those respective gene coding sequences. Post hybridization washes, once each for 15 minutes at 60–62°C, were HB, 1:1 HB:2x SSCT, 2x SSCT, 0.5x SSCT, 0.2x SSCT. Pre-antibody blocking was carried out for 30 minutes in TBST with 10% sheep serum (Sigma) and 1 mg/ml BSA. Anti-Dig, anti-Fl or anti-DNP antibodies (1:500–1000 dilution) were incubated for 1hr in TBST with 5% sheep serum and 0.1mg/ml BSA. The staining solutions were supplemented with 2% dimethylformamide and Tween-20 was omitted during INT (brown) staining. Red staining was obtained with FastRed tablet sets (Sigma F-4648) supplemented with 50mM MgCl2. A detailed version of the WMISH protocol is available upon request.
Morpholino substituted antisense oligonucleotides
Morpholino substituted antisense oligonucleotides (MOs) used here were provided by GeneTools, LLC. They were stored and injected as described previously (Ransick and Davidson, 2006)). The gcm-MO [5′-GCTTTGGAGTAACCTTCTG CACCAT-3′] was previously described. Six1-MO [5′-CCCAGGTCCGTGGCAAGGATAGGAT-3′], injected at 250µM, is a translation blocking-MO targeting a portion of the 5′ leader sequence established using 5′ RACE and PCR analyses. The confirmed translation start site does not correspond to the Spsix1/2 gene model SPU_17379. An updated six1 coding sequence has been filed in GenBank [accession JQ264781]. To confirm the efficacy of six1-MO, a fragment of the six1::gfp that contained the Gfp coding sequence fused immediately downstream of the target sequence was TA-cloned into pGEM-T easy vector (Promega), and capped mRNA for injection was transcribed with T7 Message Machine (Ambion). No Gfp was detectable from this mRNA when co-injected with six1-MO at 1:1000 ratio. c-rel-MO [5′-CGATCATTCAATCTACCTTGATAGT-3′], injected at 200–300µM, targets the confirmed sequence at the exon three to intron three splice junction. This target was chosen after 5′ RACE and PCR analyses confirmed that the gene structure upstream of exon 3 differed significantly from the Spc-rel gene model, SPU_12203. The efficacy of c-rel-MO was determined by a conventional PCR assay using primers spanning the targeted splice junction. cDNA made from embryos injected with 300µM c-rel-MO, generated a prominent amplicon for the incorrectly spliced c-rel mRNA variant and no amplicon representing the correctly spliced mRNA.
Results
Identification of late acting module
A previous cis-regulatory analysis of gcm, focused on integration of the DN signaling through suppressor-of-hairless (SuH) inputs (Ransick and Davidson, 2006). That study made use of alignments between genome sequence for Strongylocentrotus purpuratus (Sp) and sequence for a Lytechinus variegatus (Lv) BAC spanning the gcm gene (Brown et al., 2002; Brown et al., 2005). Three regions of relatively high sequence conservation were identified, then PCR-cloned and used to construct modular reporter constructs that recapitulated the early expression [e.g. DE-Sp-P-gfp]. The essential early expression enhancer (early module, E) contains SuH sites that integrate DN signaling, while the distal (D) and proximal (P) modules confer endodermal repression and basal promoter functionalities, respectively.
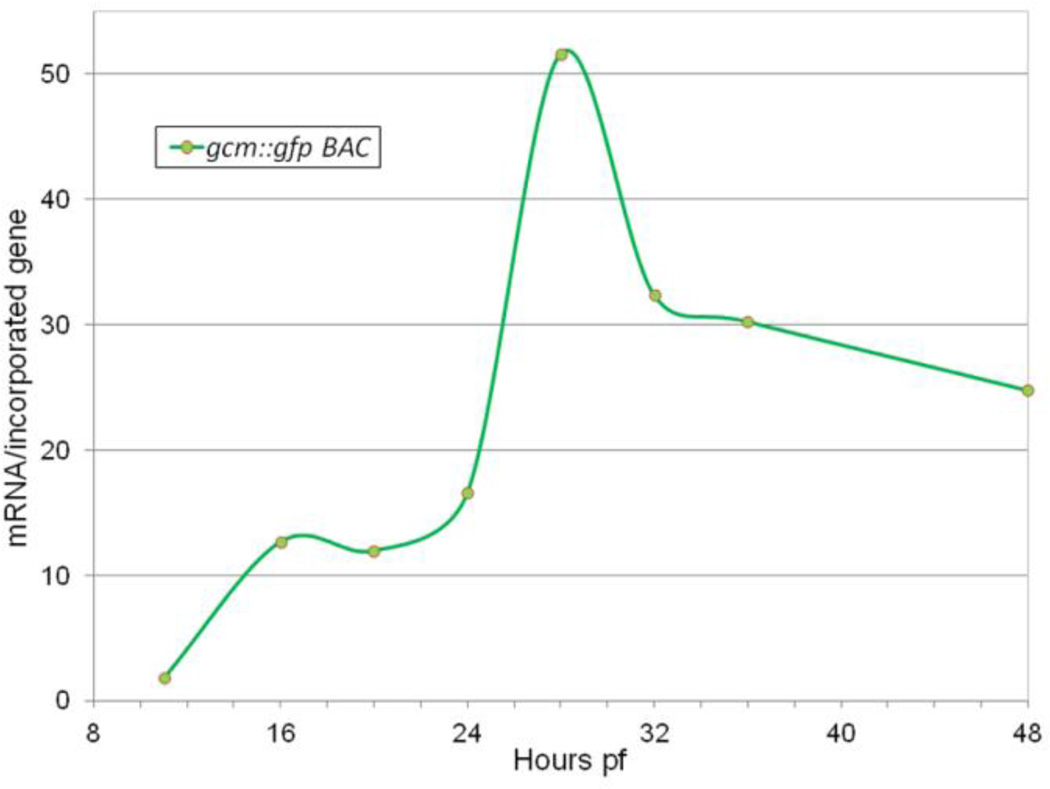
However, the early module reporter D-E-Sp-P-gfp does not ramp up at the early mesenchyme blastula, like either the endogenous gene (Fig. 1A) or an injected gcm recombinant BAC, gcm::gfp (Fig. 2). In fact, gfp output from early module constructs weakens during the mesenchyme blastula stage (Fig. 3A) and is not detectable in pigment cell precursors post-ingression. On the other hand, gcm::gfp has persistent expression in differentiated pigment cells (Fig. 3B). This confirms the expectation that regulatory elements responsible for driving late expression are present in this gcm BAC.
Fig. 2.
Graphic representation of recombinant gcm BAC (gcm::gfp) expression profile as measured by real time PCR measurement of gfp mRNA output (mRNA per incorporated gene) in 11–48hpf injected embryos; see Materials and methods for normalization details
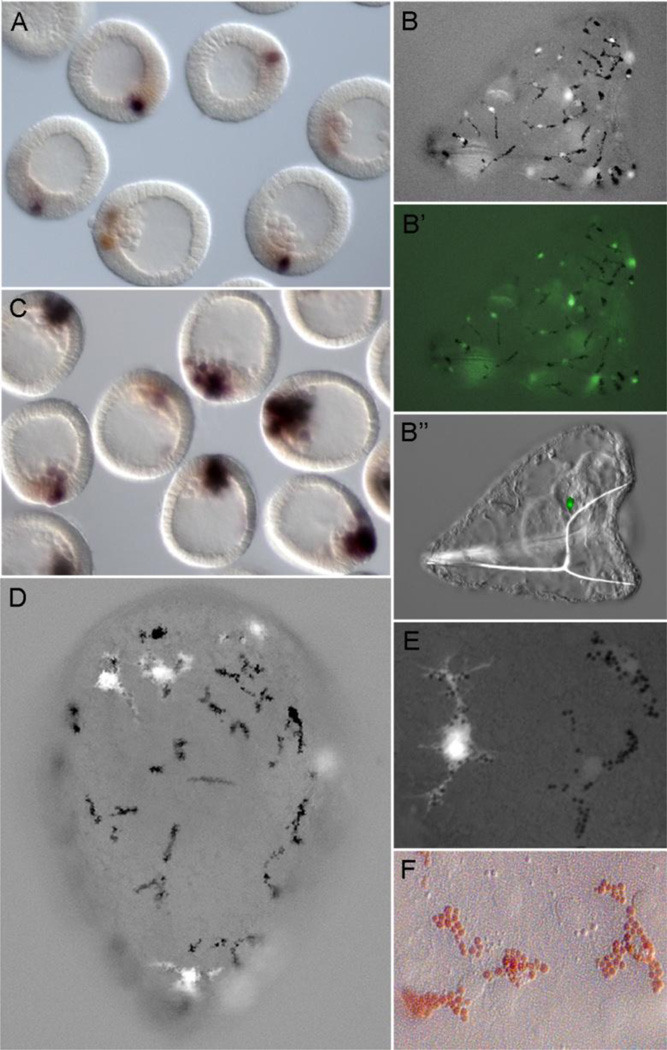
Fig. 3.
Late module expression. A) gfp output is weak by early mesenchyme blastula measured by WMISH for injected early module reporter D-E-Sp-P-gfp (blue-black); endogenous gcm (brown) B) Pluteus larva (~72hpf) that developed after injection of gcm::gfp recombinant BAC with pigment cells containing nuclearized-Gfp (B, B’) and a Gfp+ cell near the coelomic rudiment (B”). C, D, E) Expression of the late module reporter, D-G-P-gfp, is robust in pigment cell precursors at late mesenchyme blastula (C) and in differentiated pigment cells (D, E). Note the finely branched filopodia of differentiated pigment cells that become evident with cytoplasmic Gfp expression. F) Differentiated pigment cells in situ, reveals the prominent red echinochrome granules and a central nucleus; close apposition with aboral ectoderm is also evident from many ectodermal nuclei in same focal plane. Imaging conditions in B, D, E: low level transmitted light, plus Gfp epifluoresence filter set.
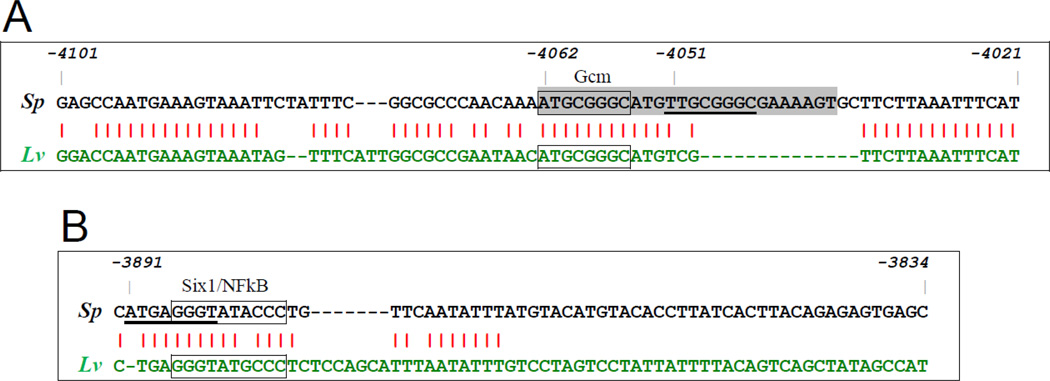
Further in silico analysis of gcm BAC 33-O18 was guided by knowledge of the effects of injecting gcm-MO, which knocks down gcm mRNA levels at least three-fold at 24hpf (Ransick and Davidson, 2006). These results are consistent with feedback circuitry, such as auto-regulation or a stabilizing intergenic loop with a gcm target gene (Davidson et al., 2003; Davidson and Levine, 2008). A search for potential Gcm binding sites in gcm BAC 33-O18, which extends from −17.6kb to +33.4kb relative to gcm transcription start, yielded a single site [ATGCGGGC] starting at position −4062. This candidate Gcm site matched the published consensus ATGCGGRY (after De Iaco et al., 2006) and was perfectly conserved in aligned Lv sequence. No additional complete matches to the consensus were found in the genomic sequence extending to position −66kb, where the next gene is located. A detailed Sp/Lv sequence comparison in the vicinity of the Gcm site match at −4062, using the alignment software Family Relations II in Dot Plot View (Brown et al., 2005), revealed this site lies in a cluster of relatively short conserved elements spanning about 500 bases (Fig. S2). A genomic fragment 512 bases in length [−4269 to −3757], designated in shorthand as G module, was PCR amplified and used to build new reporter constructs in combination with the D and P modules, e.g. D-G-P-gfp.
When injected into embryos, G module constructs consistently produced low reporter output prior to PMC ingression, yet showed increasing Gfp levels during the mesenchyme blastula stage (Fig. 3C), then maintained robust output in differentiated pigment cells (Figs. 3D, E). Importantly, the onset of robust output from G module reporter constructs in early mesenchyme blastula temporally coincides with the onset of increased output from the endogenous gene (phase two in Fig. 1A), suggesting this region as a good candidate for cis-regulatory analysis of the driver(s) of this late phase expression. However, the output from G module reporters continues to increase for several hours beyond when the endogenous gene achieves peak expression and is down regulated. This non-correspondence is likely linked to the high copy number of the injected transgene, but could also reveal a repressive modulator not present in G module.
We note two additional sequence elements within Sp G module that are partial matches to the Gcm consensus sequence. These include the tTGCGGGC element starting at −4051 with a mismatch in position 1 (Fig. 4A) and the ATGaGGGT element starting at −3891 with a mismatch in position 4 (Fig. 4B). As the corresponding regions of Lv sequence entirely lacks sequence aligning to the −4051 element and the Lv sequence aligning to the −3891 element [cTGaGGGT] has mismatches in two critical (i.e. invariant) bases, it raises doubts as to whether either of these Sp partial Gcm sites are functional. There is also a 6/8 matching element in D module [ATcgGGGT; at −13376] with mismatches at positions 3 and 4. However, this site is clearly not functional since D+/G− reporters (e.g. D-P-gfp or D-E-Sp-P-gfp) do not drive late expression.
Fig. 4.
Alignments of S. purpuratus (Sp) and L. variegatus (Lv) genomic sequence near critical driver elements in the late module. A) Interspecies sequence conservation is high in the vicinity of the 8/8 Gcm consensus site match at −4062 (box), but low near the 7/8 element at −4051 (underlined); gray highlighting indicates 26 base segment deleted in Gdel1 mutants. B) Sequence of the Sp G59 fragment and corresponding Lv region showing no sequence conservation in the downstream half, but high conservation around a palindromic element (box). This region contains prospective Six1 and NFκB sites, and partially overlaps a 7/8 match to a Gcm site (underlined).
Cis-perturbations targeting the late module
The two possible autoregulatory sites at −4062 and −4051 in G module were removed by deletion of a 26 base pair element [−4062 to −4037] to create the D-Gdel1-P-gfp construct. This construct produced less output as measured by a variety of assays. WMISH for gfp mRNA in D-Gdel1-P-gfp injected embryos revealed relatively weak signal and smaller clone sizes beginning at late mesenchyme blastula stage (Fig. 5A versus 5D). Likewise, the fluorescent protein reporter level in the pigment cells of older embryos was distinctly weaker from D-Gdel1-P-gfp than from a co-injected reporter with intact late module, D-G-P-rfp (Fig. 5E versus 5F). Finally, real time PCR quantitation of gfp mRNA levels further clarified the critical timeframe of the autoregulatory input (Fig. 6). Initially the mutated D-Gdel1-P36-gfp reporter produced a similar ascending output profile as the co-injected wild type D-G-P36-rfp. However, as the intact reporter was reaching maximal expression in the gastrula stage, the output of D-Gdel1-P36-gfp weakened. In repeated trials, the D-Gdel1-P36-gfp construct lacking Gcm sites at −4062 and −4051 did not reach the peak output level attained by the intact reporter (Table 1). And, output was maintained at about two-fold lower in late gastrula and early pluteus larva stages. These different assays consistently show Gcm sites are required for peak output from the late module. They also confirm that an auto-regulatory input contributes to maintenance of gcm expression in dispersed pigment cells.
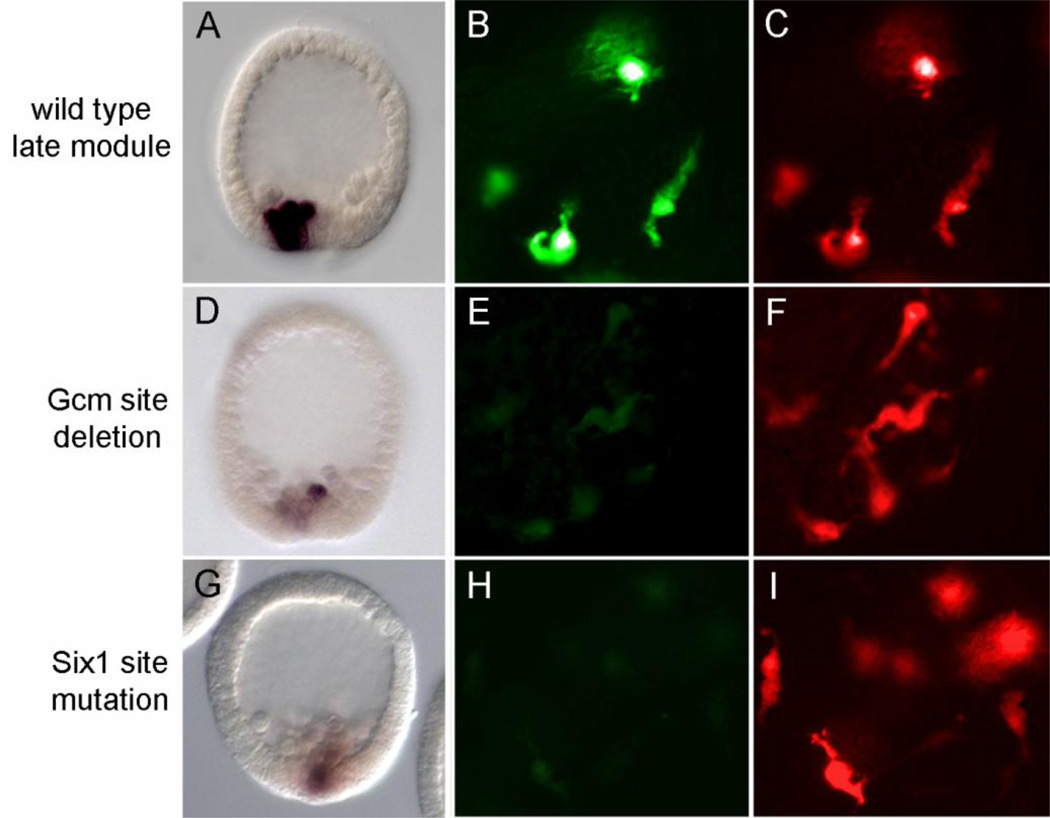
Fig. 5.
Visual assays for late module reporter output. (A, D, G) Localization of gfp mRNA by WMISH in mesenchyme blastula stage embryos; (B–C, E–F, H–I) Fluorescent protein signals (Gfp and Rfp) in pigment cells of two-day embryos. Embryos injected with the intact late module reporters D-G-P-gfp (A, B) or D-G-P-rfp (C, F, I) have strong gfp signals at late blastula and have strong Gfp or Rfp signals in pigment cells. Embryos injected with the modified late module reporters D-Gdel1-P-gfp (D, E) or D-Gm7-P-gfp (G, H) have relatively weak outputs in these assays.
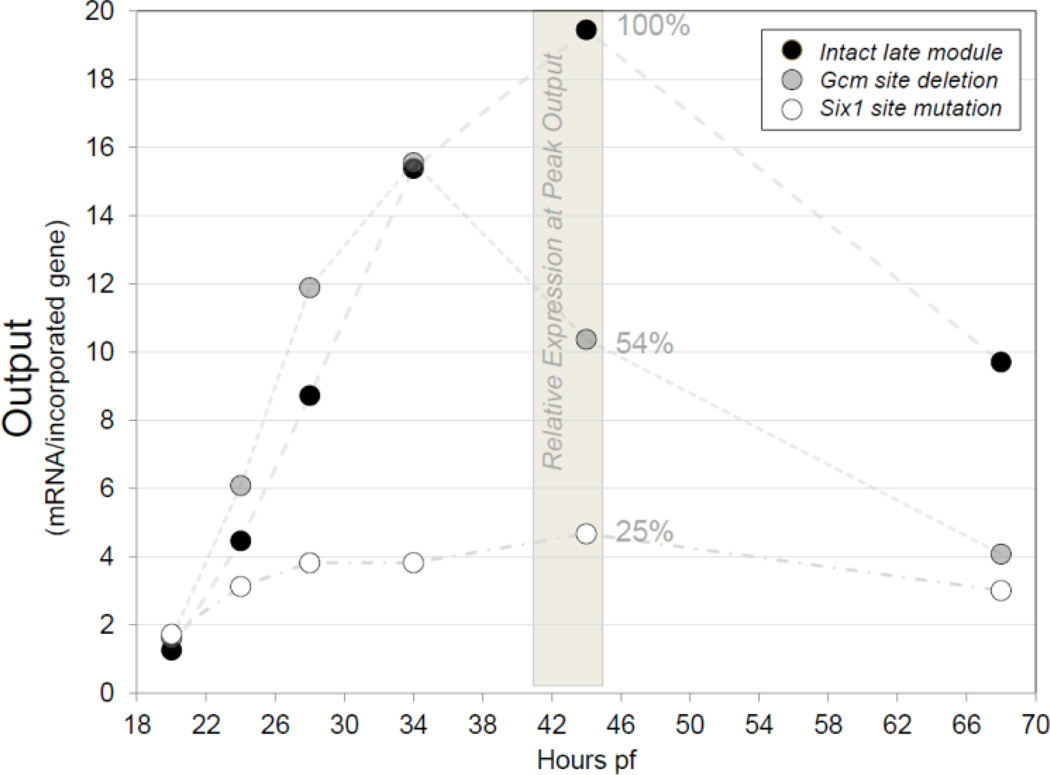
Fig. 6.
Graphic representation of real time PCR data illustrating effects of cis-perturbations on the output (mRNA/incorporated gene) of co injected late module reporters. The output profile from the intact late module reporter D-G-P36-rfp (black), sampled from 20–68hpf, serves as a highly reproducible assay of late module expression. The intact late module reporter shows a strong increase in output after PMC ingression and achieves a peak sometime during gastrulation. A time point representing Peak Output is obtained by sampling at regular intervals between 20 and 48 hrs. Here, samples from 44hpf are used for comparisons of relative expression. The output of D-Gdel1-P36-gfp (gray), the co-injected late module reporter with the Gcm site deletion, typically shows a normal initial output increase. However, relative expression is consistently weaker at Peak Output, here 54% at 44hpf. The relative expression of D-Gm7-P36-gfp (white), the late module reporter with the Six1 site mutated, is consistently low at all stages assayed, here showing a typical result with just 25% of the control Peak Output.
Table 1.
Late module relative expression at peak output
| Perturbation Type | Construct/Expression Assayed | # Expts |
Peak Output Sampled (hpf) |
Relative Expression (Avg) |
|---|---|---|---|---|
| Gcm site deleted a | 3 | 36,36,44 | 49% | |
| cis-perturbations | Six1 site mutated a | 4 | 36,37,40,44 | 16% |
| Gcm::gfp BAC, Six1 site mutated b | 1 | 28 | 40% | |
| Intact late module + Six1 MO c | 3 | 40,40,40 | 30% | |
| trans-perturbations | Gcm site del late module + Six1 MO c | 2 | 40,35 | 20% |
| Gcm mRNA + Six1 MO c | 4 | 35,40,28,30 | 36% | |
| Intact late module + gataE MO c | 2 | 32,32 | 20% |
Expression relative to:
intact late module;
intact gcm::gfp BAC;
same as shown, without MO inject
An additional driver input(s) to the late module is suggested by the normal output profile of D-Gdel1-P36-gfp throughout the mesenchyme blastula stage, which actually matches the rising output of the intact reporter D-G-P36-rfp. To localize additional driver element(s) within the late module, the 512 bp fragment was subjected to deletion analysis (Fig. S3). This led to identification of an essential 59 bp fragment (G59: −3892 to −3834), in which the only Sp/Lv sequence conservation is within the upstream 30 bases (Fig. 4B). When this 30 base sequence was checked against databases that compile binding sites for transcription factors, significant matches were identified within the 12 base palindrome region [AGGGTATACCCT] for the NFκB consensus GGGRNWYYCC (JASPAR, (Sandelin et al., 2004) and a Six1 consensus sequence, GGGTATCA (UniPROBE, (Newburger and Bulyk, 2009). The latter match was particularly noteworthy, since ongoing endomesodermal GRN perturbation analyses have identified six1 as a Gcm target knocked down sharply after gcm-MO injection or blocking DN signaling (see current EM-GRN model at http://sugp.caltech.edu/endomes/#Veg-21-30-NetworkDiagram). The G59 fragment also contains the 7/8 Gcm site match at −3891 partially overlapping with the palindromic element (Fig. 4B).
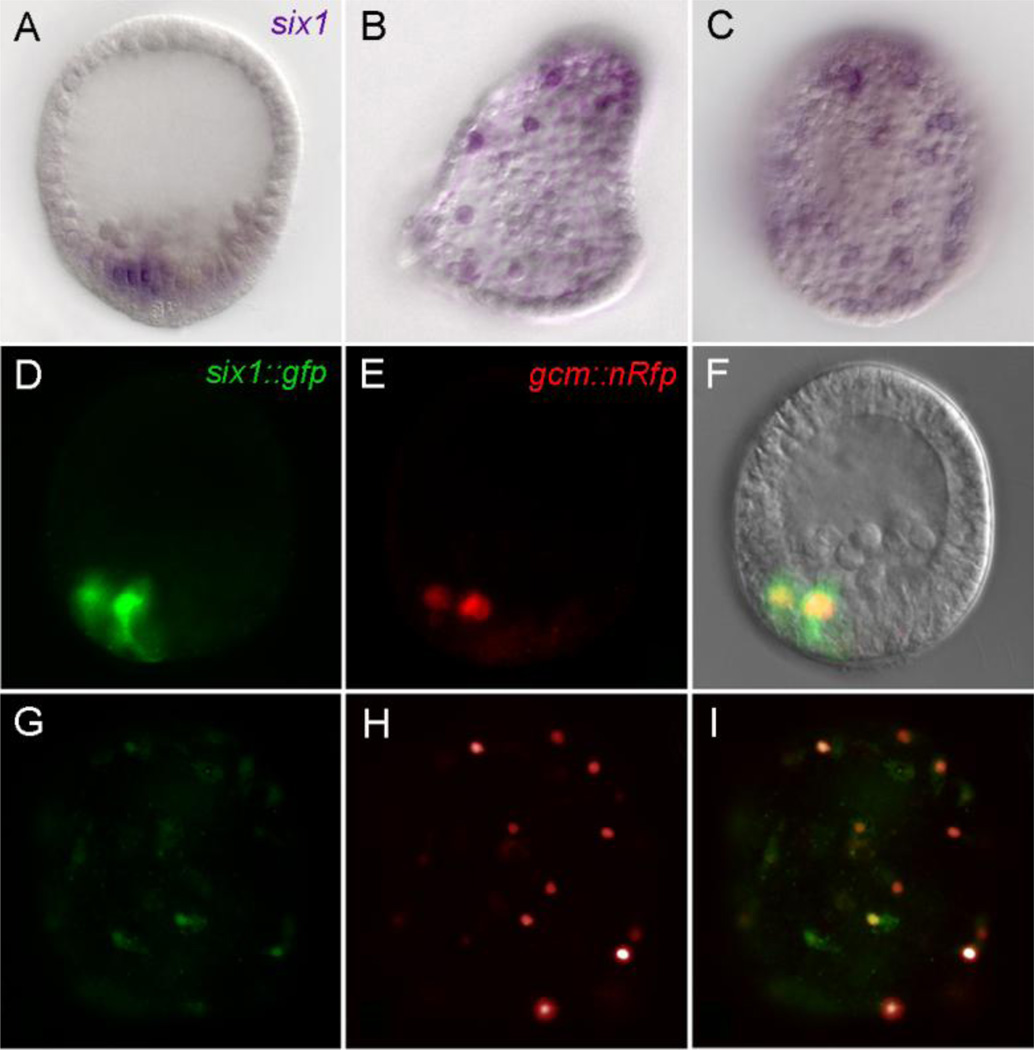
The relevance of these in silico candidates was checked by WMISH for six1, c-rel and nfkb mRNAs. Interestingly, six1 mRNA localizes to the pigment cell precursors as early as the mesenchyme blastula stage (Poustka et al., 2007) and expression is maintained in dispersed pigment cells (Figs. 7A–C). In contrast, c-rel and nfkb were enriched in dispersed pigment cells (Fig. S4), but gave no detectable WMISH signal in earlier stages. An additional confirmation that six1 is co-expressed with gcm was obtained after co-injecting a gcm reporter with recombinant six1 BAC (six1::gfp) (Figs. 7D–I).
Fig. 7.
Six1 expression localized by WMISH. (A–C) and as injected recombinant BAC six1::gfp (D, G). A) Six1 mRNA in mesenchyme blastula is confined to the aboral nonskeletogenic mesodermal domain. B, C) Six1 mRNA is enriched in scattered cells underlying the aboral ectoderm, likely to be pigment cells. D–I) Injected six1::gfp BAC (green fluorescence, D, G) co-expresses with a gcm reporter (red, E, H) in vegetal plate mesoderm at the blastula stage and in the pigment cells of two day embryos; F and I are merged images of panels shown to the left.
A mutation analysis of the conserved portion of G59 fragment showed significantly lower output from reporter constructs carrying mutations that focused on the GGGTAT portion of the palindromic element (Fig. S5, Gm6, Gm7 and Gm10). For example, the Gm7 mutation [AtttTATAaaaT] of the late module produced very low output levels: Gfp fluorescence was very weak in mature pigment cells when assayed visually (Figs. 5G–I), and real time PCR showed relative expression of just 25% of peak output of the intact reporter (Fig. 6). Output was essentially abolished from a late module reporter containing both a Gm7 mutation in addition to the Gdel1 deletion. Mutations immediately downstream of the GGGTAT element had no significant effect on gfp fluorescence output (Fig. S5 Gm11, Gm12), while mutation of sequence immediately upstream of this element (Fig. S5 Gm14) moderately affected output levels. The latter result is consistent with some autoregulatory functionality for the 7/8 Gcm site at −3891 that partially overlaps the GGGTAT element.
To further test whether the GGGTAT element in the late module is essential for late expression, a new recombinant gcm BAC (gcm::gfp del2) was made in which just the palindromic sequence containing the GGGTAT element was deleted. Embryos injected with gcm::gfp del2 showed robust Gfp expression into the mesenchyme blastula stage, which we interpret as due to the presence of an intact early module that integrates DN signaling. However, Gfp output weakened afterward, and was not detectable in the dispersed pigment cells of three-day larvae. These observations were confirmed using real time PCR quantitation of output from gcm::gfp del2 (Fig. S6). The low output of gcm::gfp del2 demonstrates that no compensatory Six1 sites exist in the gcm BAC clone. In addition, this result indicates the modest role that autoregulatory inputs play in late module output, since the Gcm sites at −4062 and −4051 are intact in gcm::gfp del2. Instead, the essential character of the G59 GGGTAT element is strongly confirmed by the reduced output of gcm::gfp del2.
Trans-perturbations targeting the late module
Six1 and c-rel mRNAs localized by WMISH to the pigment cell lineage suggest potential direct inputs that could drive second phase gcm expression. If these factors are directly involved, knockdown of their expression should produce a significant negative effect on endogenous gcm or late module reporter constructs. Morpholino antisense oligonucleotides (MOs) were designed to block six1 mRNA translation or c-rel mRNA processing. Control assays confirmed that these MOs were performing as designed and gave confidence that injected embryos would be depleted for their respective target proteins (see Methods section). Injection of 200–300µM c-rel-MO did not produce any observable phenotype nor any significant effect on endogenous gcm mRNA levels measured by real time PCR at 24 or 48hpf (data not shown). Similarly, injection of 250µM six1-MO did not produce any discernable effect on embryonic development, particularly with regard to pigment cell formation or differentiation. Significantly though, injection of 250µM six1-MO resulted in an average three-fold lower peak output of endogenous gcm in real time PCR assays (Table 1). Similarly, when late module reporters (wild type or Gcm site lacking) were co-injected with 250µM six1-MO and assayed by real time PCR, the peak output averaged three to four-fold below controls (Table 1). Thus, the portion of the gcm expression profile most affected by Six1 depletion is also that strongly affected by the cis-mutations around the candidate Six1 site. The similarity of outcomes from these cis- and trans-perturbations is consistent with a direct interaction of Six1 with this GGGTAT element in the late module. We conclude Six1 is an important regulator of gcm expression in the second phase that begins after PMC ingression.
Recent mesodermal GRN network level analyses, the details of which are to appear elsewhere, show that gataE is downstream of gcm and upstream of six1 (S. Materna, 2011). Specifically, measurement of mRNA levels of over 200 regulatory gene transcripts by Nanostring nCounter at the mesenchyme blastula stage after gataE-MO injection demonstrated that six1 is one of just two transcription factor mRNAs, the level of which is significantly depressed following knockdown of gataE transcripts (Fig. S7). As Fig. S7 demonstrates there are clearly no general off target effects of this MO. Thus, gataE-MO is useful to the current study as a perturbation reagent known to specifically deplete six1 mRNA. Indeed, when 250um gataE-MO was injected we obtained strongly reduced peak output from co-injected late module reporter D-G-P36-rfp (Table 1). This result provides independent confirmation that peak output from the gcm late module is dependent on normal six1 mRNA levels.
It is worth noting that six1-MO injection results on average in reduction of the gcm mRNA level to one third that of control peak expression in mesenchyme blastula, which is from 3600 down to ~1200 mRNA copies/embryo. Yet, six1-MO injected embryos proceed with pigment cell specification, dispersal and differentiation. This developmental outcome is quite unlike gcm-MO injected embryos, which consistently fail to specify pigment cells and so produce pigment-less ‘albino’ larvae (Ransick and Davidson, 2006). The contrasting developmental outcomes of these perturbations clarify the mesodermal GRN architecture. Clearly, six1, which is not significantly expressed before the late hatched blastula, is functioning at a level in the network that follows primary specification of pigment cell precursors.
Finally, it is likely that the recombinant gcm::gfp BAC construct when injected acted as an (unintentional) trans-perturbation reagent at the level of Gcm binding site(s). In short, the design of this knock-in inserted a gfp-SV40 polyA+ fragment into Gcm exon three, creating a 5’ in-frame fusion and resulting in production of a true Gcm-Gfp fusion protein that has the N-terminal 128 amino acids of Gcm added to the N-terminus of Gfp. Since the nuclear localization sequence as well as the DNA binding domain of Gcm is encoded within this region (Tuerk et al., 2000), output from this recombinant BAC has the potential to enter nuclei and bind to consensus Gcm sites. However, this fusion protein does not contain the more 3’ Gcm exons encoding the trans-activation domains. Therefore, within gcm::gfp expressing clones the fusion protein could compete with the endogenous protein for Gcm binding sites in gcm and other target genes, e.g. gataE or six1. In fact, we observed that gcm::gfp injected embryos show strong Gfp expression at the blastula stage, reflecting a robust reporter response to DN signaling integrated through the early module. Then after 3–5 days the resulting pluteus larvae showed a relatively weak Gfp expression in differentiated pigment cells. Gfp expression is only reliably detected here because the Gcm-Gfp fusion protein concentrates in pigment cell nuclei (e.g. see Figs. 3B, C). The difference was strikingly evident in comparison to the robust output of intact late module reporters at comparable stages (Fig. 3D). This reduced Gfp expression is likely to reflect the combined effects of the Gcm-Gfp fusion protein disrupting auto-regulation and target gene output. Importantly though, this result provides an additional corroboration that Gcm has a role in driving its own expression, especially after the late module is controlling gene output in differentiated pigment cells.
Discussion
The findings presented here extend our understanding of the cis-regulatory logic controlling gcm expression. We find that a second, later acting regulatory module becomes operational in the specified pigment cell precursors of hatched blastula stage embryos. This late module drives gcm expression to reach peak levels by the mid to late mesenchyme blastula stage and maintains output in a defined cohort of cells through the processes of ingression, dispersal and final differentiation into pigment cells.
The late module provides a positive intergenic feedback loop
The results presented here indicate that the core functionalities of this regulatory module are encoded by two inputs, Gcm itself and Six1, which is in turn downstream of gcm and gataE. The self-sustaining output provided by this cis-regulatory design is easily recognizable as another example of intergenic feedback circuitry. Our growing knowledge base of developmental gene regulatory networks (GRNs) shows this design is routinely deployed in genes that run at intermediary levels to achieve a lock down of the regulatory state (Davidson, 2009). By maintaining expression of key regulatory factors these circuits sustain the cell specification state independent of transient early embryonic patterning mechanisms. The genes on the intermediary network level in turn orchestrate expression of the differentiation gene batteries, sometimes directly or in other cases by way of an additional level of regulatory genes (Davidson, 2006). Thus the ontogeny of larval pigment cells follows from a compact network architecture: two regulatory modules controlling the gene for an essential regulatory factor. The early module, operating downstream of DN signaling as a transcriptional toggle switch, drives gcm to initially specify a founder cell population (Ransick and Davidson, 2006). A subset of the founder cells are further specified as the pigment cell precursors by maintaining a high level of gcm expression through engagement of the positive intergenic feedback loop of the late module. Finally, gcm promotes differentiation of pigment cells by driving genes in the echinochrome synthesis pathway, such as polyketide synthases and flavin-monooxygenases (Calestani et al., 2003; Calestani and Rogers, 2010). It bears repeating that this shallow regulatory hierarchy is characteristic of Type 1 embryogenesis (Davidson, 1991, 2001). These network connections are summarized in Fig. S8.
Intergenic stabilization loops so effectively couple regulatory genes into functional sets that they can persist over great evolutionary distances (Hinman et al., 2003), and this intriguing possibility should be explored in regards to the gcm /six1 feedback circuitry described here. A related goal is to establish whether this regulatory architecture is deployed whenever gcm/six1 co-expression exists in this species. The coelomic rudiment cells of pluteus larva provide a good candidate for additional study, as mRNAs for both genes have been detected there by WMISH (Poustka et al., 2007; McCauley et al., 2010). Another instance deserving additional investigation arises from the cloning of six1 from an adult coelomocyte cDNA library (Cameron et al., 2000), coupled with the enriched gcm expression in an adult coelomocyte type, the echinochrome-containing red spherule cells (J. Rast & A. Ransick, unpublished). If it can be established that these are bona fide examples of gcm/six1 co-expression, it will be revealing to ascertain the regulatory architecture deployed.
Gcm expression in relation to the mesodermal GRN
In cleavage stage embryos, the onset of expression of gcm is known to precede that of six1 by at least eight hours. Thus, there is clearly an additional cis-regulatory requirement for six1 expression that is not satisfied until the hatched blastula stage (~19hpf in Sp at 15°C). A regulatory gene that is a good candidate in this capacity is gataE, which is co-expressed with gcm in veg2 lineage a few hours after gcm activation (Lee and Davidson, 2004; Lee et al., 2007) but well in advance of six1 activation. In fact, as the connections within the mesodermal GRN have been identified, the contribution of gataE to the ontogeny of pigment cells has become increasingly apparent. Cis-regulatory analyses have shown that gataE early expression relies on DN signaling acting directly through SuH sites (Lee et al., 2007) and that the essential pigment cell differentiation gene, pks, has direct GataE inputs (Calestani and Rogers, 2010). Also noteworthy, are recent mesodermal GRN network level analyses, the details of which are to appear elsewhere (S. Materna, 2011), that show gataE is downstream of gcm and upstream of six1. Thus, the current mesodermal GRN that accounts for pigment cell specification places gcm in an intergenic feedback loop with gataE and six1. Frequent updates of relevant segments of the EM-GRN model can be viewed at http://sugp.caltech.edu/endomes/#Veg-21-30-NetworkDiagram.
An oral segment of the non-skeletogenic mesoderm territory eventually yields the plesiomorphic echinoderm mesodermal lineages for larval muscle, coelomic rudiments and blastocoelar cells. Specification starts with the emergence of a different regulatory landscape in the late hatched-blastula stage. Under the influence of Nodal expression on the oral side, processes are activated that both promote a new set of mesodermal regulatory factors, including orthologs of ese, prox, gataC and scl (Poustka et al., 2007; Rizzo et al., 2006; Duboc et al., 2010) and create a non-permissive environment for gcm. The specification mechanisms operating to carve out an oral mesoderm territory and maintain a balance between oral and aboral mesodermal fates are downstream of the TGFb-family signaling (Duboc et al., 2010). How the effectors of this pathway, such as SMAD factors or intermediate targets like the homedomain protein, Not (Materna 2011), regulate the key non-skeletogenic mesodermal transcription factors are remaining questions outside the focus of this report.
Gcm in mature pigment cells
There is still much to learn about the function of this interesting, early differentiating mesodermal lineage of echinoids. There is a growing consensus of opinion that the mesenchymal cells of echinoderm larvae (blastocoelar and pigment cells in sea urchins) have immune cell functionalities (Smith, 2005; Furukawa et al., 2009). We are therefore intrigued that the critical element that mediates Six1 interactions with the late module overlaps an NFκB consensus site match. Although our investigation found no evidence that NFκB factors play a role in regulating gcm during the specification processes of early embryogenesis, WMISH shows that c-rel and nfkb mRNAs are present in young larva in mesenchymal cells that are likely to include the pigment cells (echinochrome leeches out during the WMISH protocol) (Fig. S4). This presents the intriguing possibility that mature pigment cells are poised to respond to NFκB family signaling. Looking forward, the implication that this well known inflammatory response pathway modulates larval mesenchyme cell behavior, whether through gcm or other genes, lends support to the argument that these cells possess immune functionalities.
Highlights.
new cis-regulatory control mechanisms of glial cells missing in sea urchin embryos.
late acting cis-regulatory module of gcm becomes active in mesenchyme blastulae.
late module encoding drives gcm in a stabilizing intergenic loop with six1.
sustained gcm output locks down pigment cell fate.
late module continues to drive gcm in differentiated pigment cells.
Supplementary Material
Fig. S1 High density expression profile of gcm from unfertilized egg through 48hpf (data from Materna et al., 2010).
Fig. S2 Sp/Lv conserved elements in the vicinity of the GCM consensus site. Captured screen shots of Family Relations II in Dot Plot View with search criteria set to identify every 10 nt element with 90% conservation (red spots) in the selected regions of Sp BAC 33-O18 and Lv BAC 18-J3. A) Plot of all conserved elements within ~2800 nt segment centered on the GCM consensus site. B) Effectively zooming in, by restricting the analysis to a central 800 nt segment (blue box in A & B), reveals more clearly the cluster of conserved elements surrounding the GCM site (blue spot).
Fig. S3 Schematic diagram summarizing various deletions within the late module and their effects on reporter expression in pigment cells; all constructs were identical outside of the late module; Gcm site (blue triangle); Six1 site (yellow triangle). Ultimately, a series of 3’ deletions (G–J) narrowed down the location of a critical element to just 59 bases (G59). Mutations targeting the Six1 site of G59 similarly extinguish late module output.
Fig. S4 c-rel mRNA localized by WMISH in post-gastrula embryos to mesenchymal cells distributed under aboral ectoderm, likely to represent expression in pigment cells.
Fig. S5 Summary of mutation analysis of the conserved portion of G59 fragment. Mutations targeting the upstream GGGTAT portion of the palindromic element (blue highlighting) strongly reduced output of G module reporters.
Fig. S6 Graphic representation of expression profiles for the wild type gcm BAC, gcm::gfp (solid line) compared to gcm::gfp del2 (dashed line), which differed only in having the Six1 site deleted. Expression measured by real time PCR; plotted as gfp mRNA output (mRNA per incorporated gene) in 20–48 hpf injected embryos.
Fig. S7 Evaluation of gataE-MO injections (from Materna, 2011). RNA was extracted from injected embryos and quantified using the Nanostring nCounter. Counts for RNA from perturbed embryos are plotted against counts from unperturbed embryos. Genes that are not affected by the perturbation align on the diagonal. The small number of genes affected by gataE-MO indicates that the morpholino is specific and does not cause any broad developmental defects. Dotted lines indicate a threshold of two-fold change. Importantly, gataE-MO results in about 3 fold lower Six1 level.
Fig. S8 Current GRN model for non-skeletogenic mesoderm. Complete endomesodermal GRN model can be viewed at http://sugp.caltech.edu/endomes/#Veg-21-30-NetworkDiagram
Acknowledgments
We are pleased to acknowledge Stefan Materna for helpful discussions and for sharing his mesodermal GRN data prior to its publication; Julie Hahn and Ping Dong for BAC constructs, and the labs of S. Fraser, E. Rothenberg and B. Wold for sharing resources. This work is supported by NIH grant HD-37105.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arnone MI, Dmochowski IJ, Gache C. Using reporter genes to study cis-regulatory elements. In: Ettensohn CA, Wessel GM, Wray GA, editors. Development of Sea Urchins, Ascidians, and Other Invertebrate Deuterostomes: Experimental Approaches, Methods Cell Biol. vol. 74. Elsevier; 2004. pp. 621–652. [DOI] [PubMed] [Google Scholar]
- Brown CT, Rust AG, Clarke PJ, Pan Z, Schilstra MJ, De Buysscher T, Griffin G, Wold BJ, Cameron RA, Davidson EH, Bolouri H. New computational approaches for analysis of cis-regulatory networks. Dev. Biol. 2002;246:86–102. doi: 10.1006/dbio.2002.0619. [DOI] [PubMed] [Google Scholar]
- Brown CT, Xie Y, Davidson EH, Cameron RA. Paircomp, FamilyRelationsII and Cartwheel: tools for interspecific sequence comparison. BMC Bioinformatics. 2005;6:70. doi: 10.1186/1471-2105-6-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calestani C, Rast JP, Davidson EH. Isolation of pigment cell specific genes in the sea urchin embryo by differential macroarray screening. Development. 2003;130:4587–4596. doi: 10.1242/dev.00647. [DOI] [PubMed] [Google Scholar]
- Calestani C, Rogers DJ. Cis-regulatory analysis of the sea urchin pigment cell gene polyketide synthase. Dev. Biol. 2010;340:249–255. doi: 10.1016/j.ydbio.2010.01.026. [DOI] [PubMed] [Google Scholar]
- Cameron RA, Mahairas G, Rast JP, Martinez P, Biondi TR, Swartzell S, Wallace JC, Poustka AJ, Livingston BT, Wray GA, Ettensohn CA, Lehrach H, Britten RJ, Davidson EH, Hood L. A sea urchin genome project: sequence scan, virtual, map, and additional resources. Proc. Natl. Acad. Sci. USA. 2000;97:9514–9518. doi: 10.1073/pnas.160261897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce JC, McClay DR. Dynamics of Delta/Notch signaling on endomesoderm segregation in the sea urchin embryo. Development. 2010;137:83–91. doi: 10.1242/dev.044149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH. Spatial mechanisms of gene regulation in metazoan embryos. Development. 1991;113:1–26. doi: 10.1242/dev.113.1.1. [DOI] [PubMed] [Google Scholar]
- Davidson EH. Gene Regulatory Networks in Development and Evolution. Academic Press/Elsevier; San Diego: 2006. The Regulatory Genome. [Google Scholar]
- Davidson EH. Network design principles from the sea urchin embryo. Curr. Opin. Genet. Dev. 2009;19:535–540. doi: 10.1016/j.gde.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH, Levine MS. Properties of developmental gene regulatory networks. Proc. Natl. Acad. Sci. USA. 2008;105:20063–20066. doi: 10.1073/pnas.0806007105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH, McClay DR, Hood L. Regulatory gene networks and the properties of the developmental process. Proc. Natl. Acad. Sci. USA. 2003;100:1475–1480. doi: 10.1073/pnas.0437746100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Iaco R, Soustelle L, Kammerer M, Sorrentino S, Jacques C, Giangrande A. Huckebein-mediated autoregulation of Glide/Gcm triggers glia specification. EMBO J. 2006;25:244–254. doi: 10.1038/sj.emboj.7600907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboc V, Lapraz F, Saudemont A, Bessodes N, Mekpoh F, Haillot E, Quirin M, Lepage T. Nodal and BMP2/4 pattern the mesoderm and endoderm during development of the sea urchin embryo. Development. 2010;137:223–235. doi: 10.1242/dev.042531. [DOI] [PubMed] [Google Scholar]
- Furukawa R, Takahashi Y, Nakajima Y, Dan-Sohkawa M, Kaneko H. Defense system by mesenchyme cells in bipinnaria larvae of the starfish, Asterina pectinifera. Dev. Comp. Immunol. 2009;33:205–215. doi: 10.1016/j.dci.2008.08.011. [DOI] [PubMed] [Google Scholar]
- Gibson AW, Burke RD. The origin of pigment cells in embryos of the sea urchin Strongylocentrotus purpuratus. Dev. Biol. 1985;107:414–419. doi: 10.1016/0012-1606(85)90323-9. [DOI] [PubMed] [Google Scholar]
- Gibson AW, Burke RD. Migratory and invasive behavior of pigment cells in normal and animalized sea urchin embryos. Exp. Cell Res. 1987;173:546–557. doi: 10.1016/0014-4827(87)90294-1. [DOI] [PubMed] [Google Scholar]
- Hinman VF, Nguyen AT, Cameron RA, Davidson EH. Developmental gene regulatory network architecture across 500 million years of echinoderm evolution. Proc. Natl. Acad. Sci. USA. 2003;100:13356–13361. doi: 10.1073/pnas.2235868100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PY, Davidson EH. Expression of Spgatae, the Strongylocentrotus purpuratus ortholog of vertebrate GATA4/5/6 factors. Gene Expr. Patterns. 2004;5:161–165. doi: 10.1016/j.modgep.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Lee PY, Nam J, Davidson EH. Exclusive developmental functions of gatae cis-regulatory modules in the Strongylocentrorus purpuratus embryo. Dev. Biol. 2007;307:434–445. doi: 10.1016/j.ydbio.2007.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Materna SC, Nam J, Davidson EH. High accuracy, high-resolution prevalence measurement for the majority of locally expressed regulatory genes in early sea urchin development. Gene Expr. Patterns. 2010;10:177–184. doi: 10.1016/j.gep.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Materna SC. Ph.D. Thesis. Pasadena, CA: California Institute of Technology; 2011. The regulatory origin of oral and aboral mesoderm in sea urchin embryos. [Google Scholar]
- McCauley BS, Weideman EP, Hinman VF. A conserved gene regulatory network subcircuit drives different developmental fates in the vegetal pole of highly divergent echinoderm embryos. Dev. Biol. 2010;340:200–208. doi: 10.1016/j.ydbio.2009.11.020. [DOI] [PubMed] [Google Scholar]
- McMahon AP, Flytzanis CN, Hough-Evans BR, Katula KS, Britten RJ, Davidson EH. Introduction of cloned DNA into sea urchin egg cytoplasm: replication and persistence during embryogenesis. Dev. Biol. 1985;108:420–430. doi: 10.1016/0012-1606(85)90045-4. [DOI] [PubMed] [Google Scholar]
- Minokawa T, Rast JP, Arenas-Mena C, Franco CB, Davidson EH. Expression patterns of four different regulatory genes that function during sea urchin development. Gene Expr. Patterns. 2004;4:449–456. doi: 10.1016/j.modgep.2004.01.009. [DOI] [PubMed] [Google Scholar]
- Newburger DE, Bulyk ML. UniPROBE: an online database of protein binding microarray data on protein-DNA interactions. Nucleic Acids Res. 2009;37:D77–D82. doi: 10.1093/nar/gkn660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveri P, Carrick DM, Davidson EH. A regulatory gene network that directs micromere specification in the sea urchin embryo. Dev. Biol. 2002;246:209–228. doi: 10.1006/dbio.2002.0627. [DOI] [PubMed] [Google Scholar]
- Oliveri P, Davidson EH. Gene Regulatory Network Analysis in Sea Urchin Embryos. In: Ettensohn CA, Wessel GM, Wray GA, editors. Development of Sea Urchins, Ascidians, and Other Invertebrate Deuterostomes: Experimental Approaches, Methods In Cell Biol. vol. 74. Elsevier; 2004. pp. 775–794. [DOI] [PubMed] [Google Scholar]
- Oliveri P, Tu Q, Davidson EH. Global regulatory logic for specification of an embryonic cell lineage. Proc Natl Acad Sci U S A. 2008;105:5955–5962. doi: 10.1073/pnas.0711220105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poustka AJ, Kuhn A, Groth D, Weise V, Yaguchi S, Burke RD, Herwig R, Lehrach H, Panopoulou G. A global view of gene expression in lithium and zinc treated sea urchin embryos: new components of gene regulatory networks. Genome Biol. 2007;8:R85. doi: 10.1186/gb-2007-8-5-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransick A. Detection of mRNA by in situ hybridization and RT-PCR. In: Ettensohn CA, Wessel GM, Wray GA, editors. Development of Sea Urchins, Ascidians, and Other Invertebrate Deuterostomes: Experimental Approaches, Methods In Cell Biol. vol. 74. Elsevier; 2004. pp. 601–620. [Google Scholar]
- Ransick A, Davidson EH. cis-regulatory processing of Notch signaling input to the sea urchin glial cells missing gene during mesoderm specification. Dev. Biol. 2006;297:587–602. doi: 10.1016/j.ydbio.2006.05.037. [DOI] [PubMed] [Google Scholar]
- Ransick A, Rast JP, Minokawa T, Calestani C, Davidson EH. New early zygotic regulators expressed in endomesoderm of sea urchin embryos discovered by differential array hybridization. Dev. Biol. 2002;246:132–147. doi: 10.1006/dbio.2002.0607. [DOI] [PubMed] [Google Scholar]
- Revilla-i-Domingo R, Minokawa T, Davidson EH. R11: a cis-regulatory node of the sea urchin embryo gene network that controls early expression of SpDelta in micromeres. Dev. Biol. 2004;274:438–451. doi: 10.1016/j.ydbio.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Rizzo F, Fernandez-Serra M, Squarzoni P, Archimandritis A, Arnone MI. Identification and developmental expression of the ets gene family in the sea urchin (Strongylocentrotus purpuratus) Dev. Biol. 2006;300:35–48. doi: 10.1016/j.ydbio.2006.08.012. [DOI] [PubMed] [Google Scholar]
- Sandelin A, Alkema W, Engstrom P, Wasserman WW, Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Sherwood DR, McClay DR. LvNotch signaling mediates secondary mesenchyme specification in the sea urchin embryo. Development. 1999;126:1703–1713. doi: 10.1242/dev.126.8.1703. [DOI] [PubMed] [Google Scholar]
- Smith LC. Host responses to bacteria: innate immunity in invertebrates. In: McFall-Ngai M, Ruby BHN, editors. Advances in Molecular and Cellular Microbiology. Vol. 10. Cambridge Univ. Press; 2005. pp. 293–320. [Google Scholar]
- Sweet HC, Gehring M, Ettensohn CA. LvDelta is a mesoderm-inducing signal in the sea urchin embryo and can endow blastomeres with organizer-like properties. Development. 2002;129:1945–1955. doi: 10.1242/dev.129.8.1945. [DOI] [PubMed] [Google Scholar]
- Takata H, Kominami T. Behavior of pigment cells closely correlates the manner of gastrulation in sea urchin embryos. Zoolog. Sci. 2004;21:1025–1035. doi: 10.2108/zsj.21.1025. [DOI] [PubMed] [Google Scholar]
- Tuerk EE, Schreiber J, Wegner M. Protein stability and domain topology determine the transcriptional activity of the mammalian glial cells missing homolog, GCMb. J. Biol. Chem. 2000;275:4774–4782. doi: 10.1074/jbc.275.7.4774. [DOI] [PubMed] [Google Scholar]
- Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 2005;33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 High density expression profile of gcm from unfertilized egg through 48hpf (data from Materna et al., 2010).
Fig. S2 Sp/Lv conserved elements in the vicinity of the GCM consensus site. Captured screen shots of Family Relations II in Dot Plot View with search criteria set to identify every 10 nt element with 90% conservation (red spots) in the selected regions of Sp BAC 33-O18 and Lv BAC 18-J3. A) Plot of all conserved elements within ~2800 nt segment centered on the GCM consensus site. B) Effectively zooming in, by restricting the analysis to a central 800 nt segment (blue box in A & B), reveals more clearly the cluster of conserved elements surrounding the GCM site (blue spot).
Fig. S3 Schematic diagram summarizing various deletions within the late module and their effects on reporter expression in pigment cells; all constructs were identical outside of the late module; Gcm site (blue triangle); Six1 site (yellow triangle). Ultimately, a series of 3’ deletions (G–J) narrowed down the location of a critical element to just 59 bases (G59). Mutations targeting the Six1 site of G59 similarly extinguish late module output.
Fig. S4 c-rel mRNA localized by WMISH in post-gastrula embryos to mesenchymal cells distributed under aboral ectoderm, likely to represent expression in pigment cells.
Fig. S5 Summary of mutation analysis of the conserved portion of G59 fragment. Mutations targeting the upstream GGGTAT portion of the palindromic element (blue highlighting) strongly reduced output of G module reporters.
Fig. S6 Graphic representation of expression profiles for the wild type gcm BAC, gcm::gfp (solid line) compared to gcm::gfp del2 (dashed line), which differed only in having the Six1 site deleted. Expression measured by real time PCR; plotted as gfp mRNA output (mRNA per incorporated gene) in 20–48 hpf injected embryos.
Fig. S7 Evaluation of gataE-MO injections (from Materna, 2011). RNA was extracted from injected embryos and quantified using the Nanostring nCounter. Counts for RNA from perturbed embryos are plotted against counts from unperturbed embryos. Genes that are not affected by the perturbation align on the diagonal. The small number of genes affected by gataE-MO indicates that the morpholino is specific and does not cause any broad developmental defects. Dotted lines indicate a threshold of two-fold change. Importantly, gataE-MO results in about 3 fold lower Six1 level.
Fig. S8 Current GRN model for non-skeletogenic mesoderm. Complete endomesodermal GRN model can be viewed at http://sugp.caltech.edu/endomes/#Veg-21-30-NetworkDiagram