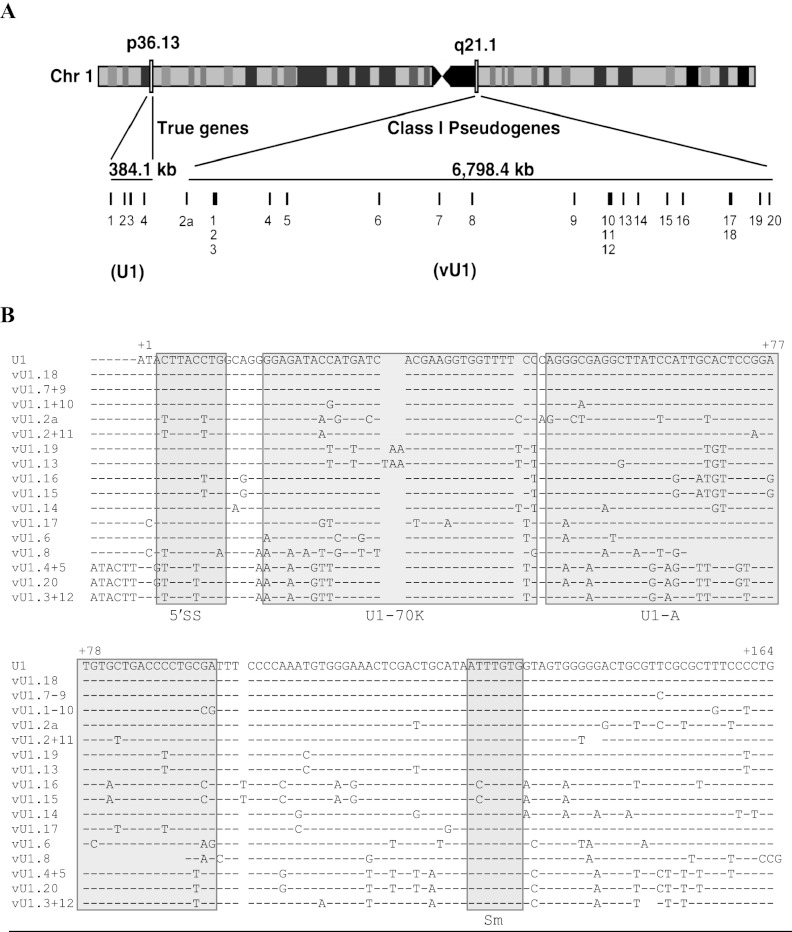
Figure 1.
Annotation and sequence of the U1 snRNA class I pseudogenes. (A) Schematic of chromosome 1 illustrating the location of U1 snRNA genes (1p36) and the U1 snRNA class I pseudogenes (1q12-21). The size of each locus is shown in kilobases (kb). Each gene is represented by a vertical line and a number. Light and dark boxes on chromosome 1 refer to regions of high and low gene activity, respectively. (B) Alignment of the snRNA sequence of the U1 snRNA genes (U1.1–4) with the 21 vU1 snRNA genes. Paired vU1 snRNAs have identical sequences. Nonconserved bases are denoted with the letter code representing one of the four nucleotide bases. A dash indicates the presence of a corresponding base, and a gap indicates the absence of a base at that position. The bases are numbered, beginning with 1 for the first base of the U1 snRNA sequence. Important features of the U1 snRNA are boxed and indicated underneath the alignment: 5′ splice site recognition motif (5′ss), U1-70K protein binding region (U1-70K); U1-A protein binding region (U1-A); Sm binding site (Sm).