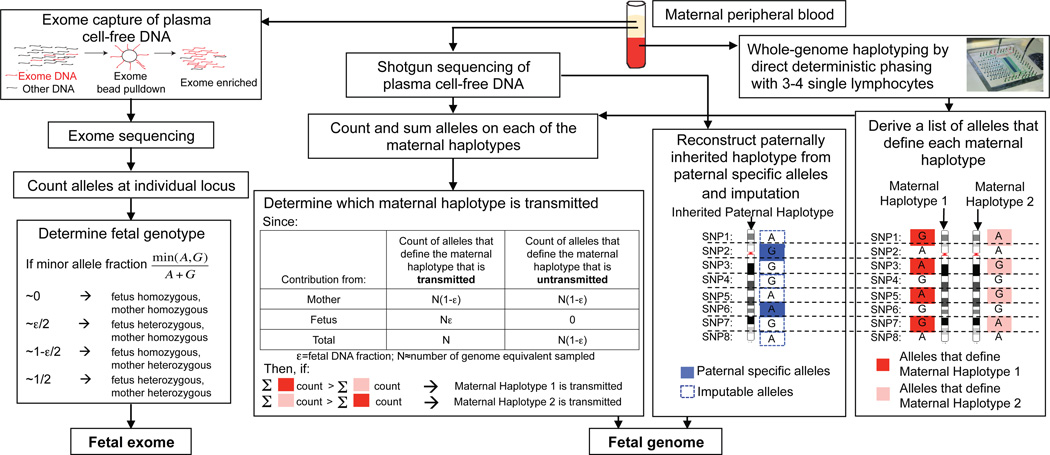
Figure 1.
Molecular counting strategies for measuring the fetal genome noninvasively from maternal blood only. Genome-wide, chromosome length haplotypes of the mother are obtained using direct deterministic phasing. The inheritance of maternal haplotypes is revealed by sequencing maternal plasma DNA and summing the count of the alleles specific to each haplotype at heterozygous loci and determining the relative representation of the two alleles. The inherited paternal haplotypes are defined by the paternal specific alleles (i.e. those that are different from the maternal ones at positions where the mother is homozygous). The allelic identity at loci linked to the paternal specific alleles on the paternal haplotype can be imputed. Alternatively, molecular counting can be applied directly to count alleles at individual locus to determine fetal genotypes via targeted deep sequencing, such as exome enriched sequencing of maternal plasma DNA. For illustrative purpose, each locus is biallelic and carries the ‘A’ or ‘G’ alleles.