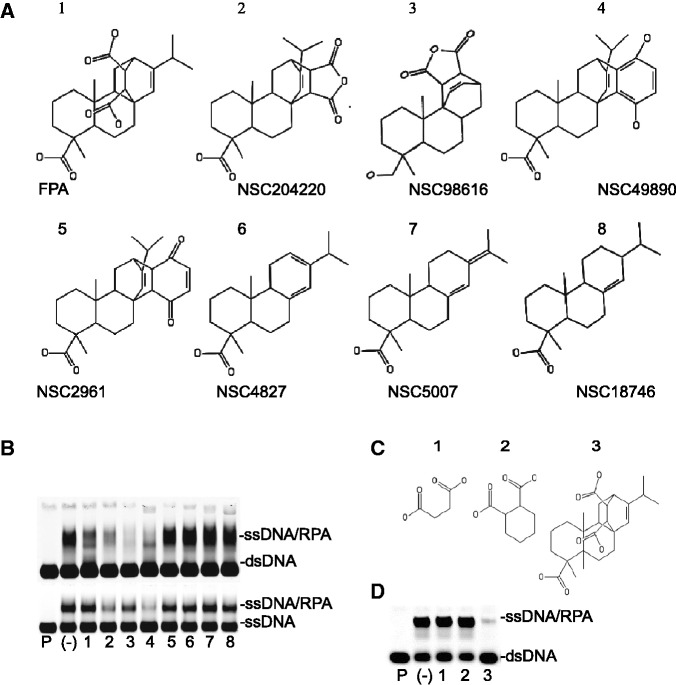
Figure 3.
Effect of structural analogues of FPA on DBD-FMBP inhibition. (A) Structures of lead compound FPA and structural analogues used in this study. (B) Effect of FPA-related compounds on DNA binding as measured by EMSAs. 200 µM of each compound was used. Top panel: addition of dsDNA to compounds preincubated with RPA. Bottom panel: addition of ssDNA to compounds preincubated with RPA. Lane with DNA in the absence of RPA is demarked by ‘P’. RPA–DNA binding without inhibitor is marked by a (-). Numbers correspond to structures in ‘A’. (C) Structure of FPA sub-structures succinic acid, 1,4-cyclohexanedicarboxylic acid and FPA (1–3, respectively). (D) Effect of compounds in ‘C’ (200 µM) on helix destabilization via EMSA. Lane with ssDNA probe in the absence of RPA is demarked by ‘P’. RPA–DNA binding without inhibitor is denoted by a (-).