Abstract
We describe a new, broadly applicable methodology for screening in parallel interactions of RNA-binding proteins (RBPs) with large numbers of microRNA (miRNA) precursors and for determining their affinities in native form in the presence of cellular factors. The assays aim at identifying pre-miRNAs that are potentially affected by the selected RBP during their biogenesis. The assays are carried out in microtiter plates and use chemiluminescent readouts. Detection of bound RBPs is achieved by protein or tag-specific antibodies allowing crude cell lysates to be used as a source of RBP. We selected 70 pre-miRNAs with phylogenetically conserved loop regions and 25 precursors of other well-characterized miRNAs for chemical synthesis in 3′-biotinylated form. An equivalent set in unmodified form served as inhibitors in affinity determinations. By testing three RBPs known to regulate miRNA biogenesis on this set of pre-miRNAs, we demonstrate that Lin28 and hnRNP A1 from cell lysates or as recombinant protein domains recognize preferentially precursors of the let-7 family, and that KSRP binds strongly to pre-miR-1-2.
INTRODUCTION
In the past years, several reports have highlighted the roles of individual RNA-binding proteins (RBP) in the regulation of microRNA (miRNA) biogenesis (reviewed in 1–3). The stem-loop structures of miRNA precursors exhibit many characteristic features that are recognized by RBPs. Examples of such features are the two-nucleotide (nt) overhangs at the 3′-ends for PAZ domain containing RBPs and the double-stranded regions in the stem of primary-miRNAs (pri-miRNAs) for nuclear factor 90 (4). Exportin 5 requires the entire stem and the 3′-overhangs to bind and exert its function (5). A further important element offering binding sites for RBPs and opportunities for control of processing are the terminal loop regions of miRNA precursors, which vary in length between 14 and 40 nts. Although short terminal loop regions can form stable structures, the longer loops may have properties akin to those of single-stranded RNAs. One of the best characterized examples is the let-7 family that bears large loop regions. These contain short conserved motifs that allow Lin28 and other RBPs to bind and regulate let-7 biogenesis (6–11). Interestingly, for a sizable fraction of miRNA precursors, terminal loop regions are highly conserved (12) presumably to enable control of miRNA precursor processing by cognate RBPs. This regulation can be stimulatory or inhibitory, it can involve individual or subsets of miRNAs and it can be nuclear or cytoplasmic (13). Many of these RBPs have been known previously for different functions such as splicing and regulation of mRNA stability. Thus, hnRNP A1 enhances Drosha cleavage of pri-mir-18a (12,14), but it retards processing of pri-let-7a-1 (15). KSRP, a mediator of mRNA decay, binds miRNA terminal loops and enhances Drosha as well as Dicer processing (16). The splicing factor SF2/ASF has been shown to affect the expression of 40 miRNAs (17), and RBM3, a cold inducible RBP, affects Dicer processing of a majority of miRNAs (18), whereas the hnRNP-like TDP-43 promotes Dicer and Drosha processing (19). These examples paint a picture of widespread control of miRNA activity via RBP-mediated regulation of their precursors.
To expound this area, new methods are needed to identify the interaction of RBPs with pri- or pre-miRNAs, to define their binding sites and affinities. We therefore elaborated a strategy of testing RBP binding in vitro as a predictor for potential effects in vivo. Here, we demonstrate a directed approach for testing pre-selected RBPs for binding to large numbers of synthetic miRNA precursors in parallel. We chemically synthesized a unique set of pre-miRNAs with highly conserved terminal loop regions (12) that are, owing to their biogenesis, also present on primary transcripts. For the assays, we chose a sensitive chemiluminescent system on microtiter plates that allowed detection of either the RNA or the protein component. We illustrate the potential of these assays to reveal and characterize both known and new interactions between 95 pre-miRNAs and three RBPs, Lin28, hnRNP A1 and KSRP, in native form and in the presence of cellular factors. These RBPs have been previously shown to bind to miRNA precursors. We show that Lin28 and hnRNP A1 select pre-let-7 family members with remarkable preference within our set of pre-miRNAs and determine the affinities of selected pre-miRNAs to hnRNP A1 by competition versions of these assays. For KSRP, we find that this protein binds prominently to pre-miR-1-2. The results suggest that the approach of screening for direct binding between RBPs and pre-miRNAs in vitro has the potential of identifying interactions that affect miRNA biogenesis.
MATERIALS AND METHODS
Synthesis of pre-miRNAs
Pre-miR sequences were extracted from information provided in reference (20) and the miRBase database (21) (http://www.mirbase.org/). The assembly of pre-miRNAs was carried out in 96-well plate format on a Dr. Oligo synthesizer (Biolytics Lab Performance, Inc.) from controlled pore glass (CPG) solid support (1000 Å with a typical loading of 35 μmol/g) and 2′-O-[(triisopropylsilyl)oxy]methyl (TOM)-protected ribonucleoside phosphoramidites. Biotin modifications at the 3′-end of the sequences were introduced by using Biotin Serinol CPG (Glen Research). For sequence assembly, a standard coupling cycle was used using 4% dichloroacetic acid in dichloromethane for the detritylation steps, 0.07 M amidite and 0.25 M ethylthiotetrazole in acetonitrile for the coupling steps, a 1:1 mixture of 20% N-methylimidazole in tetrahydrofuran and acetic acid anhydride/pyridine/tetrahydrofuran (8:1:1) for the capping steps and 0.01 M iodine in pyridine/water/acetonitrile (3:15:32) for the oxidation steps. All pre-miRNAs were produced conserving the dimethoxytrityl group at the 5′-end of the sequence. Deprotection and cleavage from the solid support were carried out in the gas phase in a pressure-proof vessel using gaseous monomethylamine for 60 min at 27 psi and 70°C. After gas phase deprotection, the RNA sequences were recovered from the solid support by treatment with triethylamine trihydrofluoride in dimethylsulfoxide/1-methyl-2-pyrrolidone/triethylamine (11:4:5), and deprotection of the 2′-O-TOM-protecting group was completed by heating the resulting solution at 65°C for 2 h. The reaction was quenched by diluting the deprotection solution with 1 M aqueous triethylammonium acetate. Purification was carried out on DNA Top-10 cartridges (Varian, Inc.). Cartridges were conditioned with acetonitrile followed by 1 M aqueous triethylammonium acetate. Subsequently, the deprotection solution containing the RNA sequences was applied onto the cartridges followed by treatment with a 3:2 mixture of 100 g/L aqueous sodium chloride and methanol. The 5′-dimethoxytrityl group of the full length product was removed by treating the column with aqueous 2% trifluoroacetic acid followed by extensive washing with water. The full-length product was recovered from the cartridge by eluting with 50 mM tetraethylammonium carbonate in acetonitrile/water (1:1). Sequences were quantified by ultraviolet absorption measurements, adjusted to a yield of 10 nmol per well and dried down.
Synthesis of oligoribonucleotides
The hnRNP A1 ‘winner’ sequences (22) 5′-UAUGAUAGGGACUUAGGGUG-3′ and a biotinylated version, 5′-UAUGAUAGGGACUUAGGGUGTTT-biotin-3′, were synthesized on a MerMade 12 instrument (Bioautomation Corporation, Plano, USA) under standard conditions. UnySupport CPG and 3′-BiotinTEG-CPG were obtained from Glen Research (Sterling, USA) and phosphoramidites from Thermo Fisher Scientific (Milwaukee, USA). RNAs were deprotected with gaseous monomethylamine (65°C at 1 bar pressure for 2 h). RNAs were eluted with EtOH/water (1:1). Desilylation was carried out on dry RNAs using freshly prepared 1-methyl-2-pyrrolidone, triethylamine (TEA) and TEA.3HF (6:3:4) for 90 min at 70°C. Isopropoxytrimethylsilane was added, and samples were lyophilized. The crude RNA was subjected to HPLC (Agilent 1200 Series; Agilent Technologies, Santa Clara, USA). Dimethoxytrityl groups were cleaved using 40% acetic acid for 30 min at RT. RNAs were again purified by HPLC using a C18 column (XBridge OST, particle size 2.5 µm; Waters, Milford, USA). Purified samples were analysed on an Agilent 6130 Series Quadrupole LC/MS (Agilent Technologies, Santa Clara, USA) with electron spray ionization. Purity and yields were determined by HPLC and Nanodrop, respectively.
Screening with directly coated recombinant proteins
White microtiter plates (96- or 384-well type, NUNC, Maxisorp) were used in all plate assays. The volumes for the 96-well format were 50 μl, and for the 384-well format, 20 μl. All washing steps were done 3–8 times with running deionized water, which allowed rapid and thorough washing. Coating with Lin28 zinc finger (ZnF) domain was carried out overnight at 5 μg/ml in 10 mM Hepes pH 7.2, 100 mM KCl, 0.5 mM DTT, 1 μM ZnCl2. The plate was emptied and left in blocking buffer (coating buffer supplemented with10 μg/ml heparin, 0.05% Tween 20, 0.1 mM DTT) for 1 h. Following a washing step with water, biotinylated pre-miRNAs were added at a concentration of 5 nM in blocking buffer for 2 h at 4°C. The plate was emptied (without washing) and exposed to a formaldehyde solution [0.5% in phosphate buffered saline (PBS)] for 5 min. This step was intended to prevent dissociation of the RNA from the RBP during subsequent steps. Formaldehyde is known to crosslink RNA to proteins (23) and to denature RNA, a capacity exploited in denaturing gel electrophoresis (24). In our experiments, the fixation step stabilized the interaction and was retained in all of our microtiter plate experiments. In the case of Lin28, signals increased by ∼50% by this measure. The fixation step may not always be necessary as demonstrated in an assay that also used directly adsorbed proteins (25). For hnRNP A1 (UP1), the coating buffer was PBS, and the blocking and binding buffer composed of 25 mM Hepes, 300 mM NaCl, 0.05% Tween 20, 0.5 mM TCEP [tris(2-carboxyethyl)phosphine hydrochloride, neutralized, Pierce, Cat. no. 77720] and 1% Top-Block (a derivative of gelatin, LuBioScience, Lucerne, Switzerland). Bound biotinylated RNA was detected by a peroxidase conjugate of streptavidin (Sigma, 0.1 μg/ml in 150 mM NaCl, 25 mM Hepes pH 7.2, 0.05% Tween 20, 1% Top-Block). The plate was again washed, and BM chemiluminescent substrate (Roche Applied Science, Cat. no. 11 582 950 001) was added. Plates were sealed with a transparent foil before measurement (26) in a luminometer (Mithras 940, Berthold, Regensdorf, Switzerland). For presentation of the data, we subtracted a general background corresponding to the average of the 5th or 10th percentiles and re-scaled the signals by dividing by the maximal values in the assay. This transformation does not change the overall shape of the bar graph. The background in wells without protein coating was 2000–3000 counts/s, whereas signals reached up to 5 000 000 counts/s.
Screening immobilized biotinylated pre-miRNAs with cell lysates
Plates were coated with streptavidin (Sigma, Cat. no. 85878 or S4762) at 2 μg/ml in PBS for 16 h or longer at 4°C and blocked with 150 mM NaCl, 25 mM Hepes pH 7.2, 0.05% Tween 20, 1% Top-Block. Biotinylated oligoribonucleotides were diluted to 4–10 nM in 25 mM Hepes pH 7.2 and allowed to bind for 3 h at room temperature or overnight at 4°C. The wells were washed with cold water and cell lysates (diluted from frozen aliquots into a buffer of 300 mM NaCl, 25 mM Hepes pH 7.2, 0.05% Tween 20, 1% Top-Block, 0.5 mM TCEP, for Lin28 additionally 10µM ZnCl2) added for 1 h at 4°C, a temperature chosen to reduce potential RNAse activity in the cell lysate. We noticed that lysates stored for prolonged periods even at −80°C showed reduced activity, particularly for Lin28. Lysate dilutions were 100 times for Lin28 and hnRNP A1 experiments, and 20 times for KSRP. The plate was emptied and fixed with formaldehyde as described earlier in the text. The bound protein of interest was detected with antibodies against Myc (clone 9E10, Santa Cruz) or anti-hnRNP A1 [clone 4B10, Santa Cruz, Cat. no. sc-32301, originally described in reference (27)]; it does not recognize hnRNP A2 or hnRNP B proteins, both at 0.1 μg/ml for 1 h at room temperature. Anti-KSRP (Bethyl Laboratories, LuBioScience, Cat. no. A302-021A) was used at 0.1 μg/mL with a 3 h incubation at 4°C. The results of western blots analysis carried out with these antibodies on Hela cell extracts were in agreement with the expected molecular weights for hnRNP A1 and KSRP (cf. Supplementary Figure S1). The buffer for primary and secondary peroxidase conjugated antibodies was 150 mM NaCl, 25 mM Hepes pH 7.2, 0.05% Tween 20, 1% Top-Block. The peroxidase conjugates against mouse and rabbit IgG (Cat. no. 074-1806 and 074-1506, respectively, KPL, BioConcept AG, Allschwil, Switzerland) were applied at a dilution of 1/3000 for 1 h. As mentioned earlier, a chemiluminescent substrate was used for signal generation. Examples of lysate dilution curves are presented in Supplementary Figure S2.
Determination of affinities by ELISA inhibition assays
For determining binding affinities, a two-step assay was used. Varying concentrations of the oligoribonucleotides to be tested were incubated with a constant dilution (1/100) of Hela cell lysate in a buffer containing 250 mM NaCl, 25 mM Hepes pH 7.2, 0.5 mM TCEP, 0.05% Tween 20 and 1% Top-Block. These mixtures, prepared in polypropylene 96-well plates (NUNC, Cat. no. 732-2620), were kept at 4°C for 3 h and, in the cold room, 20 μl aliquots transferred to white 384-well microtiter plates that had been coated for 4 h with 20 μl of 5 nM solution of a biotinylated oligoribonucleotide known to bind hnRNP A1 (UAUGAUAGGGACUUAGGGUGTTT-biotin, corresponding to the ‘winner’ sequence extended with three thymidine residues and biotin). The plate was washed with cold water to minimize temperature-dependent edge effects. After 30 min incubation at 4°C, the plate was emptied (without washing), fixed and processed as described earlier for the cell lysate screening assay with immobilized biotinylated pre-miRNAs. The data were fitted to a logistic equation (Y = (Bo-BG)/(1 + ([competitor]/IC50)sl) + BG) using the Solver feature of Excel: Y was the measured chemiluminescence and taken as proportional to the free fraction in the pre-equilibration mixture, Bo corresponded to the signals without inhibitor, BG was a constant background (1.3% of the maximal signals) and IC50 and sl (slope) the parameters to be determined. The Z′-factor (28) was 0.85.
Recombinant proteins
His6-tagged Lin28 ZnF (29) was used in the assay. Myc-tagged Lin28 was prepared as described (29), except that HEK293T cells were used as recipients of the Myc-Lin28 encoding plasmid. Recombinant UP1 was purified from Escherichia coli and consisted of residues 2-196 from hnRNPA1 (Uniprot P09651). Purification was performed by Ni-NTA chromatography of an N-terminal His6-tagged UP1 followed by the cleavage of the His6-tag by TEV protease resulting in a protein with two additional glycine residues at the N-terminus before the UP1 sequence (Dr. Pierre Barraud, ETH Zurich, manuscript in preparation).
Cell culture, transfections and preparation of cell lysates
Hela (ATCC No. CCL-2, LGC, Molsheim, FR) and HEK293T (CRL-11268, LGC, Molsheim, FR) cells were cultured in Dulbecco’s modified Eagle’s medium (Gibco) containing 10% fetal bovine serum (Sigma).
Lysates of Hela cells for inhibition experiments were prepared by washing cells with PBS and collecting with a cell scraper. Following centrifugation, the cell pellet (100 μl) was suspended in 1 ml Hepes pH 7.2, 200 mM NaCl, supplemented with a protease inhibitor mix (Complete Cat. no. 11836153001, Roche Diagnostics, Switzerland, AG). The suspension was sonicated and centrifuged at 14 000 rpm in an Eppendorf centrifuge for 10 min. Glycerol (200 μl) was added, and aliquots were frozen in liquid nitrogen followed by storage at −80°C.
MiRNA precursors associated with hnRNP A1
Immunoprecipitation was carried out for three independent biological replicates with anti-hnRNP A1 mAb (Santa Cruz 4B10) or mouse serum IgG (Sigma, I5381) as negative control. Lysates of PBS-washed Hela cells were obtained by lysis of one confluent T-75 flask in 1 ml of a buffer of 50 mM HEPES, pH 7.5, 150 mM KCl, 1% NP40, 0.5 mM DTT, 0.1% SDS, 2 mM EDTA, complete protease inhibitor EDTA-free (Roche), 50 U/mL RNasin (Promega). Antibodies were adsorbed to magnetic beads (Dyna-beads® Protein G, Invitrogen, 100-03D), according to manufacturer’s instructions (10 μg antibody/0.6 mg beads). The lysates were incubated with the 0.6 mg beads at 4°C for 1 h (5 mg total protein, final volume 500 μl equivalent to 10 mg/ml). The beads were washed five times with 50 mM HEPES, pH 7.5; 150 mM KCl; 0.05% IGEPAL, 0.5 mM DTT, 1× protease inhibitor mix (Roche Applied Biosystems). Residual traces of DNA were subjected to on-bead-digestion with RNAse free recombinant DNAse (Roche Applied Biosystems) for 15 min at room temperature, according to manufacturer’s instructions. For RNA extraction, the beads were incubated in 200 µl final volume containing 200 μg recombinant proteinase K (Roche Applied Biosystems) in digestion buffer (100 mM Tris/HCl pH 7.5, 150 mM NaCl, 12.5 mM EDTA, 2% SDS) for 15 min at 65°C with shaking followed by chloroform–phenol extraction. RNA was reverse transcribed (high capacity cDNA reverse transcription kit, Life Tech) using random hexamer primers. Quantitative reverse transcriptase polymerase chain reaction (PCR) (qRT-PCR) was performed with FastStart Universal SYBR Green Master (Rox) (Roche Applied Biosystems), according to supplier’s instructions. The primers used are listed in Supplementary Table S2. Approximately 50% of hnRNPA1 was captured from the extract after incubation with antibody coated beads, as assessed by western blot comparison of equivalent amounts (5%) of bead supernatants, eluates and total lysate.
RESULTS
Components of the screening assay
For our screening assays and as tools for further characterization, we chemically synthesized a set of 95 miRNA precursors in 3′-biotinylated and unmodified forms. The length of human pre-miRNAs typically spans 50–90 nt and likely represents the limit of solid-phase RNA synthesis using readily available reagents. An example of state-of-the-art chemical RNA synthesis is described in the recent account of the synthesis of a 110-nt putative precursor RNA using a specially developed phosphoramidite (30). We used the 2′-O-TOM phosphoramidite, which is reportedly especially effective for solid-phase synthesis of long RNAs (31). Sequences selected were for the most part pre-miRNAs with phylogenetically conserved terminal loop regions as measured by the ratio of conservation scores between loop and stem regions (12), and the remainder well-characterized miRNAs expressed in cancer cell lines (Supplementary Table S1).
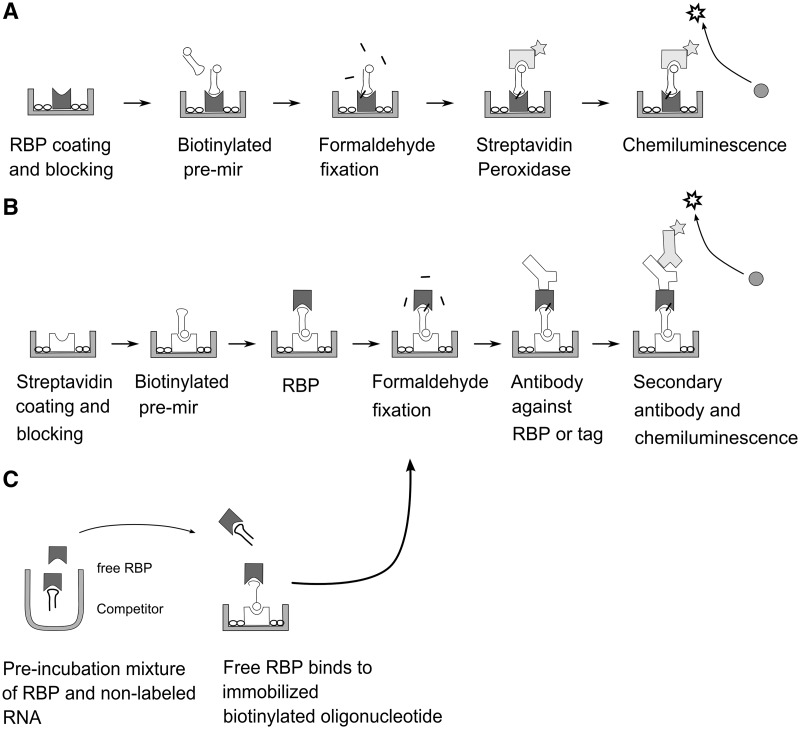
In our approach, we probed selected RBPs for binding to this set of oligoribonucleotides and used two sources of protein in the screens: recombinant purified protein in truncated forms that retain one or more RNA-binding domains and, as a second source, crude cellular lysates. During the course of our experiments and depending on the availability of detection reagents, we devised several assay configurations that are depicted and explained in the schematics and legend of Figure 1. Essentially, in the simplest configuration, we directly coated recombinant purified RBP to microtiter wells and probed with biotinylated pre-miRNA. As an alternative, we allowed RBPs to bind to immobilized biotinylated pre-miRNAs. In this case, detection of the bound protein was achieved by protein or tag-specific antibodies. A third, related assay type served to derive affinities by inhibition with unlabelled pre-miRNAs.
Figure 1.
Schematics of the three configurations used in the microtiter plate assays of this communication. (A) Binding assay with direct coating of purified RBPs (e.g. recombinant protein domains). (B) Binding assay with streptavidin immobilized pre-miRNAs and detection of RBP by antibodies. Source of RBP can be total cell lysate. (C) Inhibition assay for determining affinities of interactions. The assay is similar to that depicted in (B); the biotinylated RNA can be a short oligoribonucleotide known to bind the RBP.
Pre-let-7 family members are preferred targets of Lin28 and hnRNP A1
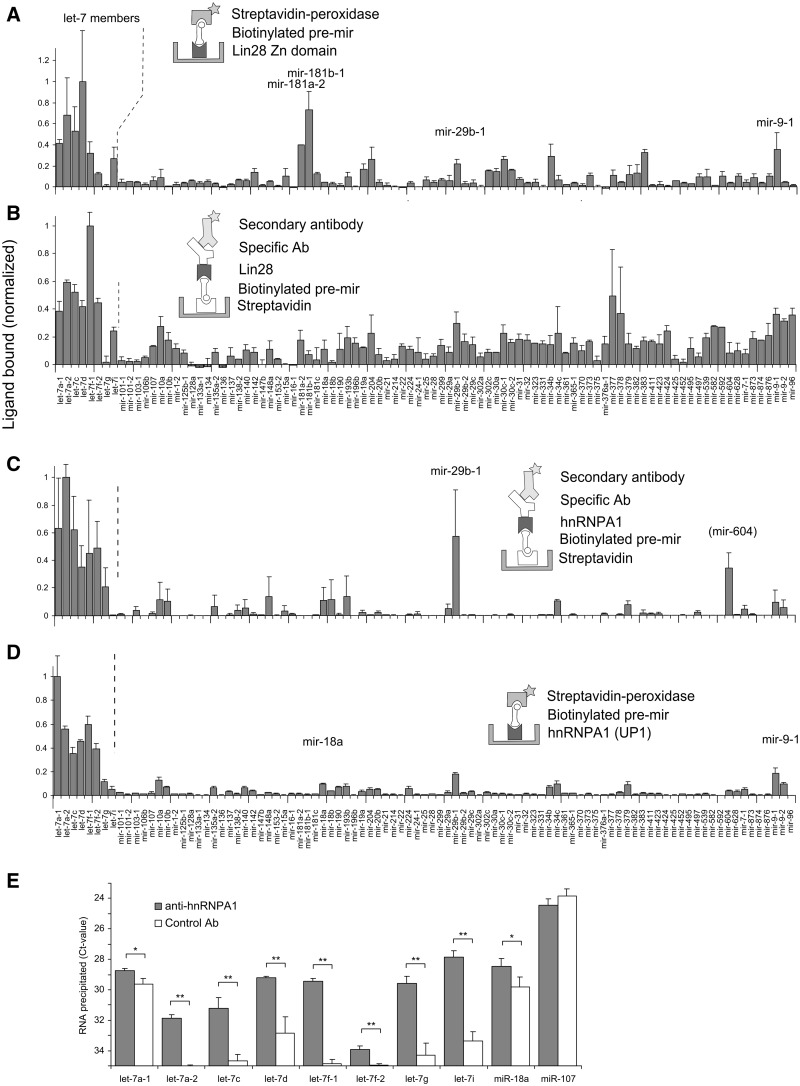
Lin28 is a small cytoplasmic RBP expressed during embryonic development. It contains an N-terminal cold shock domain (CSD) and two CCHC-type ZnF domains, all of which contribute in binding to let-7 precursors (32). The recently solved NMR structure of human Lin28 ZnF domain complexed to AGGAGAU has shown selective recognition of the pre-let-7 family members by a NGNNG consensus motif (29). We compared the binding of native Lin28 with that of the Lin28 ZnF domain, which had been used earlier to probe a small subset of this library (29) and is shown here for the full set of 95 pre-miRNAs. We adsorbed purified Lin28 ZnF domain to microtiter wells and allowed it to capture biotinylated pre-miRNAs (Figure 2A). A streptavidin-peroxidase system generated the final luminescent signals. The strong preference of Lin28 for let-7 family precursors was reinforced, though a few additional and reproducibly high-scoring precursors (miR-181 members, pre-miR-9-1) were observed. Using a reversed configuration of the assay, we then interrogated pre-bound biotinylated miRNA precursors with a lysate of cells that expressed plasmid-encoded Myc-tagged full-length Lin28 and used a tag-specific antibody to measure bound Lin28 (Figure 2B). Titrations of the cell lysates showed that dilutions in the range of 50-fold were suitable for this type of assay (Supplementary Figure S2A). Gratifyingly, the same binding events observed with the ZnF domain were present in the new profile, which again highlighted the let-7 family, miR-9-1 and miR-181 members but also contained an array of additional signals. These may have originated from the CSD or from other RBPs in the lysate that mediated Lin28 binding. Some of the signals may represent transient, non-functional contacts between the RBP and the RNA, at the terminal loop or other parts of the structure. Of note, both miR-9 and miR-181 have recently been reported to repress Lin28 expression (33,34) in a fashion similar to let-7. It remains to be clarified whether precursors of miR-9 and miR-181 bind to Lin28 in cells.
Figure 2.
Recognition of pre-miRNAs by native and recombinant RBPs in microtiter plate screening assays (A–D). The proteins assayed were as follows: (A) recombinant purified ZnF domains of Lin28 [data from a subset of the library has been presented earlier (29)]; (B) Lin28 in lysates of Myc-tagged Lin28-transfected HEK293T cells; (C) hnRNP A1 in lysates of Hela cells; (D) recombinant purified UP1 (a fragment of hnRNP A1 containing two RRM domains). Figures are representative of multiple experiments. Labels in brackets indicate signals observed in one experiment only. Error bars represent standard deviations. The labels on the X-axis of panels (A) and (C) are aligned with those of (B) and (D), respectively. The background is expressed as ratio between the average of the five lowest values in the assay and the maximal value on the plate, and it was 12.7, 7.3, 0.02 and 0.55% for screens A–D, respectively. (E) RNA-immunoprecipitation analysis of miRNA precursors associated with hnRNP A1 in Hela cells. Lysed Hela cells were incubated with anti-hnRNPA1 or control antibody coated magnetic beads. Precursor miRNAs were quantified by qRT-PCR with primers covering the pre-miRNA sequences. Averages of the cycle thresholds (Ct) from three independent biological replicates with standard deviations are shown. Asterisks denote statistical significance (two-tailed Student’s t-test) at P < 0.05 (*) and at P < 0.01 (**).
HnRNP A1 is a nucleocytoplasmic shuttling protein known for its various roles in RNA biology. It carries two copies of an RNA recognition motif (RRM) at the N-terminus and a glycine-rich domain at the C-terminus, which is responsible for cooperative binding to RNAs. hnRNP A1 also regulates Drosha processing of two miRNAs. The protein binds to pri-miR-18a, a member of the polycistronic miR-17-92 cluster (14), alters the structure of pri-miR-18a and facilitates Drosha cleavage without affecting neighbouring hairpins in the cluster. hnRNP A1 also binds to pri-let-7a-1 and acts as a repressor of let-7 biogenesis by antagonizing the docking of KSRP (15) (KH-type splicing regulatory protein), which is a component of Drosha and Dicer complexes and known to positively regulate processing of many miRNAs (16).
The human let-7 family has 10 mature miRNAs expressed from 13 distinct precursors (35). We reasoned that a simple binding screen could reveal additional hnRNP A1-regulated candidates of the let-7 precursors and other pre-miRNAs. We screened hnRNP A1 in its native form directly from Hela cell lysates using a specific antibody for detection. This antibody is specific for hnRNP A1 (27), (cf. also Supplementary Figure S1) and allowed facile detection of the protein in Hela cell lysates in this assay (Supplementary Figure S2B). Roughly one-fourth of the immobilized pre-miRNAs captured hnRNP A1 (Figure 2C). Strikingly, except for pre-let-7i, all let-7 members generated high signals. To validate these results, we reversed the format of the assay and coated the surface with UP1, a fragment of hnRNP A1 that contains the RRM domains (Figure 2D). Once again, the binding profile from a screen of the biotinylated pre-miRNAs was similar to that of the native protein from cellular lysates, indicating that the RRMs accounted for the majority of the RNA interactions seen in this assay.
Interactions of hnRNP A1 with let-7 precursors in cells
For assessing whether the interactions also occurred with endogenously expressed hnRNP A1 and miRNAs, we immunoprecipitated hnRNP A1 with antibodies from Hela cell extracts and measured the associated RNA species by qRT-PCR (Figure 2E). The primers used detected pre-miRNAs but may also recognize the respective pri-miRNAs. The enrichment factors (hnRNPA1 associated/control) spanned a range of 2-fold for let-7a-1 and let-7f-2 to levels up to 45-fold for let-7f-1 and let-7i. No enrichment was seen for the precursors of miR-107, which had been negative in the screening assays.
Affinity of hnRNP A1 against pre-miRNAs
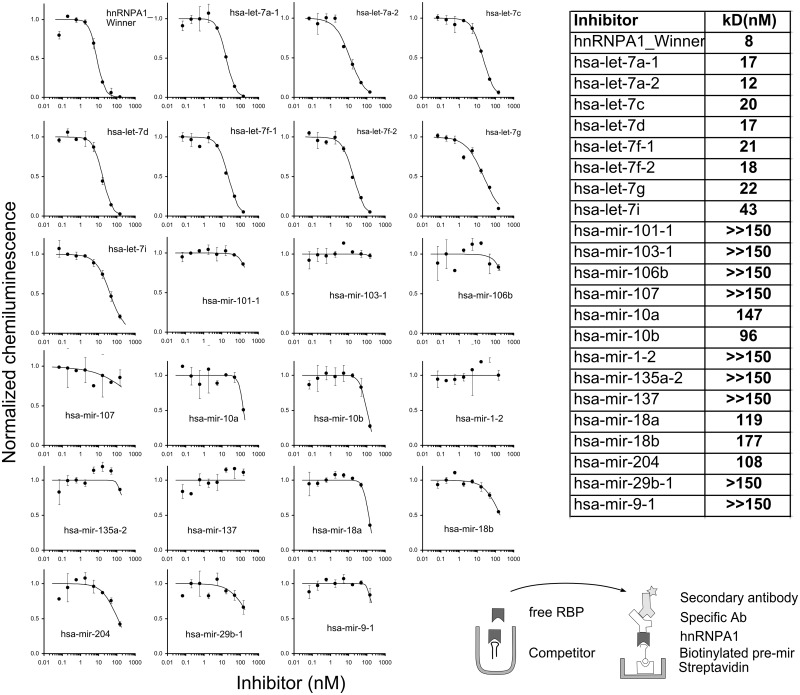
We estimated the dissociation constants for hnRNP A1 with the let-7 family members and additional pre-miRNAs by an inhibition assay, again based on RNA ELISA using captured protein as the readout (Figure 3). Importantly, the inhibition assay also served to show that the signals from the screen (Figure 2C) were owing to specific interactions between hnRNP A1 and the respective pre-miRNAs. In a pre-incubation step, we allowed graded concentrations of non-labelled pre-miRNAs to equilibrate with a constant dilution of cell lysate and, in the next step, placed these mixtures in wells that contained an immobilized biotinylated version of the SELEX winner sequence of hnRNP A1 (22). The assay was designed to measure a fraction of the free protein in the solution. Under the assumption that the equilibrium was not greatly disturbed during the time in the coated wells, we calculated IC50-values, which corresponded to the dissociation constants (Kd). For the SELEX winner sequence, we determined a Kd of 8 nM in a buffer containing 250 mM NaCl. This value is roughly in agreement with the reported Kd of 1 nM from a filter binding assay with radiolabelled RNA (22) (in a buffer of 100 mM KCl, 2.5 mM MgCl2) and a fluorescence assay with a value of 2.5 nM (36). The Kd values correlated with the binding strength observed in the screening assay (cf. Supplementary Figure S3). Pre-let-7i, which had scored negative and low in the screening assays (Figure 2C and D, respectively), displayed a measurable, but lowest ranking affinity within the pre-let-7 family. A small number of imperfections to the correlation across a large panel of 95 pre-miRNAs can be expected, especially with measurements under one set of conditions, and may arise for many reasons, including assay inherent imprecision and differences between binding to immobilized reagents compared with binding in solution. However, in a sensitive assay, such as the one of Figure 2D, even low signals seem to be meaningful.
Figure 3.
Determination of affinities of pre-miRNAs to hnRNP A1. (A) Inhibition curves for pre-miRNAs and a consensus sequence for hnRNP A1 [SELEX winner sequence (22)]. Source of hnRNP A1 was a lysate of Hela cells; pre-miRNAs were chemically re-synthesized. In this two-step assay, equilibrated mixtures of a constant lysate dilution and varying oligoribonucleotide concentrations were transferred to wells coated with biotinylated winner sequence for capturing a fraction of the non-occupied hnRNP A1. Following antibody mediated amplification and background subtraction, the signals were normalized to the non-inhibited control. Error bars indicate standard deviations of duplicates. Inset: scheme of the inhibition assay. The table shows Kd values derived from the binding curves.
In our assay, the affinities of the pre-let-7 family members spanned a range of 12–43 nM. The value for pre-let-7a-1, for which hnRNP A1 has been shown to inhibit Drosha processing by displacing KSRP, was 17 nM. Pre-miR-18a, whose processing from the pri- to the pre-form by Drosha is enhanced by hnRNP A1 (14), displayed a distinctly lower affinity of 119 nM. Possibly, the requirements for enhanced processing, where hnRNP A1 acts as an auxiliary factor for Drosha, differ from those of a merely inhibitory function.
The new observation that hnRNP A1 binds to many let-7 precursors suggests that this protein may not only displace KSRP from let-7a-1 but also from other let-7 precursors. Compared with hnRNP A1, Lin28 bound the let-7 precursors with considerably higher affinity (0.15–0.46 nM) under similar conditions (29). The screening results, together with the inhibition assay and the immunoprecipitation data, confirm the functionality of hnRNP A1-binding motifs on let-7 loop regions postulated earlier (15). In two unrelated screens of an RRM protein (to be described elsewhere), little or no binding to let-7 family members was observed, demonstrating that the screen was not biased towards this family.
KSRP binds prominently to pre-miR-1
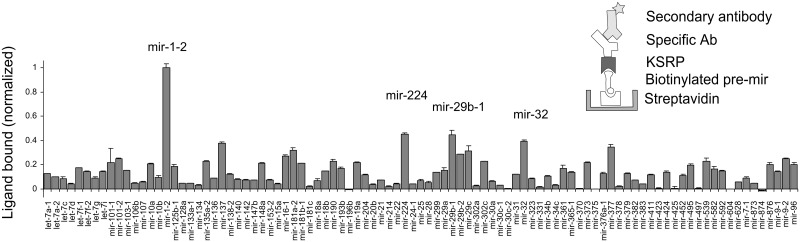
In a final example, we demonstrate the activity of KSRP in the binding screen. By western blot analysis of Hela cell lysates, the antibody proved to be specific for KSRP (Supplementary Figure S1). The screening profile (Figure 4) showed a prominent signal for pre-miR-1-2 (supported by testing at various lysate dilutions, cf. Supplementary Figure S2C) and weaker signals for various other pre-miRNAs, including pre-let-7 members. KSRP binds strongly to precursors of the muscle-specific miR-1 and supports their maturation (16). A strict correlation between KSRP’s function in the Dicer complex and its RNA binding is not expected. Still, the results of the screen support the notion of widespread, but mostly moderate augmentation of miRNA processing. Owing to its four KH domains with individually low affinity, the binding preferences of KSRP have been difficult to rationalize (37).
Figure 4.
Binding of KSRP in lysates of Hela cells to immobilized pre-miRNAs. The schematics depict the assay configuration used for the assay. Specificity for KSRP was accomplished by an antibody against the protein. One of two experiments is shown. Error bars represent standard deviations. The background, expressed as ratio between the average of the five lowest values in the assay and the maximal value on the plate, was 7.5%.
DISCUSSION
Currently, one of the major challenges in the non-coding RNA field is the elucidation of the pathways and mechanisms that regulate miRNA biogenesis. Key to this endeavour is the identification of the involved factors. Not surprisingly, the post-transcriptionally acting RBPs have turned out to play a central role in miRNA biogenesis. For a given RBP, unbiased genome-wide methods based on RNA immunoprecipitation and cross-linking such as PAR-CLIP (38) strive for a comprehensive picture for all of the coding and non-coding RNA species bound by the RBP in a selected cell type. They provide a snapshot of the interacting RNA species in a cell and also allow to identify the RNA elements recognized by RBPs. Identification of bound RNAs is performed after a multi-step protocol in which the RNA sample is converted for high-throughput sequencing. Sequence reads are subsequently sifted bioinformatically to identify interacting RNAs. A detailed mechanistic understanding of an RBP’s activity, as well as its specificity and affinity, then calls for structural and biochemical studies with the protein and its RNA partner molecules (39). In contrast to genome-wide methods, the in vitro approach of screening synthetic pre-miRNAs presented here is a system that gauges quantitatively the potential of an RBP to bind to a defined set of miRNA precursor structures in cells where both components are expressed. As this in vitro system is dissociated from the requirement of co-expression, binding of KSRP to the muscle specific pre-miR-1-2, for example, was detectable in our experiments without prior selection of the cellular system. As long as binding sites in the microtiter plates are not saturated by other RBPs, the system has the potential to detect interactions with proteins expressed with a wide range of abundances, if the sensitivity of the antibody allows. Mindful of the potential to detect transient non-functional interactions (40), a positive result implies a corresponding binding interaction in vivo and an ensuing biological effect. Our observations with the binding of Lin28 to let-7 pre-miRNAs and the interaction of KSRP with pre-miR-1-2, where the biological roles of the RBPs in miRNA biogenesis are well documented, support this concept.
An enabling aspect of our methodology is the chemical synthesis of libraries of unmodified and biotinylated pre-miRNAs. Once the collection of biotinylated pre-miRNAs is assembled, binding assays to several RBPs are quickly done and evaluated. Although the chemical synthesis of long RNAs is unable to compete in terms of yields and lengths with in vitro transcription from DNA templates, it has the advantage of allowing site-specific incorporation of functionalized nucleosides, i.e. biotin, at any desired position. Converting in vitro-transcribed RNA molecules to defined biotinylated derivatives requires considerable additional handling.
The importance of determining interaction parameters between biomacromolecules has triggered the development of numerous analytical methods. These range from qualitative binding tests to quantitative, biophysically rigorous methods such as fluorescence polarization and isothermal calorimetry that do not require separation of bound from free components. Traditionally, RNA–protein interaction studies entail radiolabelled RNAs and purified proteins, often in conjunction with gel shift or filter binding to separate bound from excess ligands. Here, we introduce related types of chemiluminescent assays that, for two versions, do not require purified RBPs nor radiolabelled components. One assay version is suitable for screening RNA–protein interactions and the second for deriving affinities. Basically, specificity for the binding protein, even in crude lysates, is achieved by an antibody specific for the protein or for a tag, if expressed in appropriate plasmids. Highly practical aspects are the ease with which assay configurations or conditions can be changed, avoidance of radioactivity and the use of the protein component for the readout. The assays with directly adsorbed recombinant proteins are technically simple and modest with respect to amounts of reagents required.
In contrast to measurements with recombinant proteins, cell lysates as a source of RBP allow probing proteins in native form, an important aspect for large or sensitive proteins and protein complexes. This type of assay also permits easily determining binding activity of an RBP from modified or drug-treated cells. We believe that the inhibition version of the assay may replace gel shift assays in many instances where RNA–protein interactions are studied and should also be suitable for testing compounds in vitro. The use of small assay volumes, as here in microtiter plates, implies that the study of RNA–protein interactions can be easily extended to a wider, possibly genomic, scale.
In this study, we screened a set of 95 pre-miRNAs in two configurations, one based on adsorbed purified recombinant protein domains and the other on immobilized miRNA precursors. For Lin28, we observed a sharper discrimination of binders with directly adsorbed proteins compared with the inverse, protein capturing assay with cell lysates. Obviously, differences can arise from the lack of RNA interaction domains, as the recombinant protein consisted of the Lin28 ZnF domains without the CSD. Less tractable differences may be owing to denaturing of proteins when directly adsorbed to surfaces and other surface phenomena. Still, both screening formats identified the let-7 family precursors in the Lin28 screen. In our second example where we compared full-length protein and a recombinant domain, the binding profiles of hnRNP A1 to pre-miRNAs were practically identical. The screen, together with the inhibition assay and supported by the RNA-immunoprecipitation experiments, revealed for the first time that hnRNP A1 interacts with many members of the let-7 family. The results confirm the functionality of hnRNP A1-binding motifs on let-7 loop regions postulated earlier (15). The affinities of a subset of pre-miRNAs measured by inhibition assay were roughly consistent with the signal intensities of the plate-binding screen. As these affinities were similar to those of pre-let-7a-1, on which hnRNP A1 is effective as a processing inhibitor, experimental testing of the hnRNP A1 effect on the other pre-let-7 members could be envisaged. It is clear that binding strength alone cannot explain or predict biological outcomes. Pre-miR-18a bound relatively poorly to hnRNP A1 in both the screening and inhibition assays. Possibly, the known cleavage-induced conformation changes reduce the affinity of hnRNP A1 to the pre-miRNA as compared with the primary transcript, which was found in co-precipitates with hnRNP A1 (14).
A variety of investigations of RBPs using these techniques is currently ongoing.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–2 and Supplementary Figures 1–3.
FUNDING
The Swiss National Science Foundation (in part) with a joint Sinergia grant [CRSII3_127454 to A.P.G. and J.H., and 205321_124720 to J.H.]. Funding for open access charge: Department of Chemistry and Applied Biosciences, ETH Zurich.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to F. Loughlin and P. Barraud for generous supply of recombinant proteins and Myc-Lin28 expressing plasmid. They thank M. Zavolan for help in retrieval of pre-miRNA sequences, M. Zimmermann for synthesis of oligonucleotides and M. Roos and L. Gebert for transfections. They also thank F. Allain and W. Filipowicz for helpful suggestions.
REFERENCES
- 1.Siomi H, Siomi MC. Posttranscriptional regulation of microRNA biogenesis in animals. Mol. Cell. 2010;38:323–332. doi: 10.1016/j.molcel.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 2.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 3.Newman MA, Hammond SM. Emerging paradigms of regulated microRNA processing. Genes Dev. 2010;24:1086–1092. doi: 10.1101/gad.1919710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sakamoto S, Aoki K, Higuchi T, Todaka H, Morisawa K, Tamaki N, Hatano E, Fukushima A, Taniguchi T, Agata Y. The NF90-NF45 complex functions as a negative regulator in the microRNA processing pathway. Mol. Cell. Biol. 2009;29:3754–3769. doi: 10.1128/MCB.01836-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okada C, Yamashita E, Lee SJ, Shibata S, Katahira J, Nakagawa A, Yoneda Y, Tsukihara T. A high-resolution structure of the pre-microRNA nuclear export machinery. Science. 2009;326:1275–1279. doi: 10.1126/science.1178705. [DOI] [PubMed] [Google Scholar]
- 6.Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor microRNA. Mol. Cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 7.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Piskounova E, Viswanathan SR, Janas M, LaPierre RJ, Daley GQ, Sliz P, Gregory RI. Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J. Biol. Chem. 2008;283:21310–21314. doi: 10.1074/jbc.C800108200. [DOI] [PubMed] [Google Scholar]
- 10.Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat. Cell Biol. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 11.Lightfoot HL, Bugaut A, Armisen J, Lehrbach NJ, Miska EA, Balasubramanian S. A LIN28-dependent structural change in pre-let-7g directly inhibits dicer processing. Biochemistry. 2011;50:7514–7521. doi: 10.1021/bi200851d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michlewski G, Guil S, Semple CA, Cáceres JF. Posttranscriptional regulation of miRNAs harboring conserved terminal loops. Mol. Cell. 2008;32:383–393. doi: 10.1016/j.molcel.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suzuki HI, Miyazono K. Emerging complexity of microRNA generation cascades. J. Biochem. 2011;149:15–25. doi: 10.1093/jb/mvq113. [DOI] [PubMed] [Google Scholar]
- 14.Guil S, Cáceres JF. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat. Struct. Mol. Biol. 2007;14:591–596. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- 15.Michlewski G, Cáceres JF. Antagonistic role of hnRNP A1 and KSRP in the regulation of let-7a biogenesis. Nat. Struct. Mol. Biol. 2010;17:1011–1018. doi: 10.1038/nsmb.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, Gherzi R, Rosenfeld MG. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu H, Sun S, Tu K, Gao Y, Xie B, Krainer AR, Zhu J. A splicing-independent function of SF2/ASF in microRNA processing. Mol. Cell. 2010;38:67–77. doi: 10.1016/j.molcel.2010.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pilotte J, Dupont-Versteegden EE, Vanderklish PW. Widespread regulation of miRNA biogenesis at the dicer step by the cold-inducible RNA-binding protein, RBM3. PLoS One. 2011;6:e28446. doi: 10.1371/journal.pone.0028446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawahara Y, Mieda-Sato A. TDP-43 Promotes microRNA biogenesis as a component of the Drosha and dicer complexes. Proc. Natl Acad. Sci. USA. 2012;109:3347–3352. doi: 10.1073/pnas.1112427109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burd CG, Dreyfuss G. RNA binding specificity of hnRNP A1: significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J. 1994;13:1197–1204. doi: 10.1002/j.1460-2075.1994.tb06369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Möller K, Rinke J, Ross A, Buddle G, Brimacombe R. The use of formaldehyde in RNA-protein cross-linking studies with ribosomal subunits from Escherichia coli. Eur. J. Biochem. 1977;76:175–187. doi: 10.1111/j.1432-1033.1977.tb11583.x. [DOI] [PubMed] [Google Scholar]
- 24.Lehrach H, Diamond D, Wozney JM, Boedtker H. RNA molecular weight determinations by gel electrophoresis under denaturing conditions, a critical re-examination. Biochemistry. 1977;16:4743–4751. doi: 10.1021/bi00640a033. [DOI] [PubMed] [Google Scholar]
- 25.King PH. RNA-binding analyses of HuC and HuD with the VEGF and c-myc 3′-untranslated regions using a novel ELISA-based assay. Nucleic Acids Res. 2000;28:e20. doi: 10.1093/nar/28.7.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Towbin H, Zingel O. Atmospheric influence on the background of luminol-based chemiluminescent assays. Anal. Biochem. 2007;369:256–257. doi: 10.1016/j.ab.2007.04.051. [DOI] [PubMed] [Google Scholar]
- 27.Piñol-Roma S, Choi YD, Matunis MJ, Dreyfuss G. Immunopurification of heterogeneous nuclear ribonucleoprotein particles reveals an assortment of RNA-binding proteins. Genes Dev. 1988;2:215–227. doi: 10.1101/gad.2.2.215. [DOI] [PubMed] [Google Scholar]
- 28.Zhang J-H, Chung TDY, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 29.Loughlin FE, Gebert LFR, Towbin H, Brunschweiger A, Hall J, Allain FH-T. Structural basis of pre-let-7 miRNA recognition by the zinc knuckles of pluripotency factor Lin28. Nat. Struct. Mol. Biol. 2012;19:84–89. doi: 10.1038/nsmb.2202. [DOI] [PubMed] [Google Scholar]
- 30.Shiba Y, Masuda H, Watanabe N, Ego T, Takagaki K, Ishiyama K, Ohgi T, Yano J. Chemical synthesis of a very long oligoribonucleotide with 2-cyanoethoxymethyl (CEM) as the 2’-O-protecting group: structural identification and biological activity of a synthetic 110mer precursor-microRNA candidate. Nucleic Acids Res. 2007;35:3287–3296. doi: 10.1093/nar/gkm202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pitsch S, Weiss PA, Jenny L, Stutz A, Wu X. Reliable chemical synthesis of oligoribonucleotides (RNA) with 2′-O-[(Triisopropylsilyl)oxy]methyl(2′-O-tom)-protected phosphoramidites. Helv. Chim. Acta. 2001;84:3773–3795. [Google Scholar]
- 32.Nam Y, Chen C, Gregory RI, Chou JJ, Sliz P. Molecular basis for interaction of let-7 microRNAs with Lin28. Cell. 2011;147:1080–1091. doi: 10.1016/j.cell.2011.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhong X, Li N, Liang S, Huang Q, Coukos G, Zhang L. Identification of MicroRNAs regulating reprogramming factor LIN28 in embryonic stem cells and cancer cells. J. Biol. Chem. 2010;285:41961–41971. doi: 10.1074/jbc.M110.169607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li X, Zhang J, Gao L, McClellan S, Finan MA, Butler TW, Owen LB, Piazza GA, Xi Y. MiR-181 mediates cell differentiation by interrupting the Lin28 and let-7 feedback circuit. Cell Death Differ. 2012;19:378–386. doi: 10.1038/cdd.2011.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18:505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 36.Abdul-Manan N, O’Malley SM, Williams KR. Origins of binding specificity of the A1 heterogeneous nuclear ribonucleoprotein. Biochemistry. 1996;35:3545–3554. doi: 10.1021/bi952298p. [DOI] [PubMed] [Google Scholar]
- 37.Briata P, Chen C-Y, Giovarelli M, Pasero M, Trabucchi M, Ramos A, Gherzi R. KSRP, many functions for a single protein. Front. Biosci. 2011;16:1787–1796. doi: 10.2741/3821. [DOI] [PubMed] [Google Scholar]
- 38.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp A-C, Munschauer M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kishore S, Jaskiewicz L, Burger L, Hausser J, Khorshid M, Zavolan M. A quantitative analysis of CLIP methods for identifying binding sites of RNA-binding proteins. Nat. Methods. 2011;8:559–564. doi: 10.1038/nmeth.1608. [DOI] [PubMed] [Google Scholar]
- 40.Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M, Ascano M, Jr, Tuschl T, Ohler U, Keene JD. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol. Cell. 2011;43:327–339. doi: 10.1016/j.molcel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.