Abstract
Nitrated polycyclic aromatic hydrocarbons are common environmental pollutants, of which many are mutagenic and carcinogenic. 1-Nitropyrene is the most abundant nitrated polycyclic aromatic hydrocarbon, which causes DNA damage and is carcinogenic in experimental animals. Error-prone translesion synthesis of 1-nitropyrene–derived DNA lesions generates mutations that likely play a role in the etiology of cancer. Here, we report two crystal structures of the human Y-family DNA polymerase iota complexed with the major 1-nitropyrene DNA lesion at the insertion stage, incorporating either dCTP or dATP nucleotide opposite the lesion. Polι maintains the adduct in its active site in two distinct conformations. dCTP forms a Watson–Crick base pair with the adducted guanine and excludes the pyrene ring from the helical DNA, which inhibits replication beyond the lesion. By contrast, the mismatched dATP stacks above the pyrene ring that is intercalated in the helix and achieves a productive conformation for misincorporation. The intra-helical bulky pyrene mimics a base pair in the active site and facilitates adenine misincorporation. By structure-based mutagenesis, we show that the restrictive active site of human polη prevents the intra-helical conformation and A-base misinsertions. This work provides one of the molecular mechanisms for G to T transversions, a signature mutation in human lung cancer.
INTRODUCTION
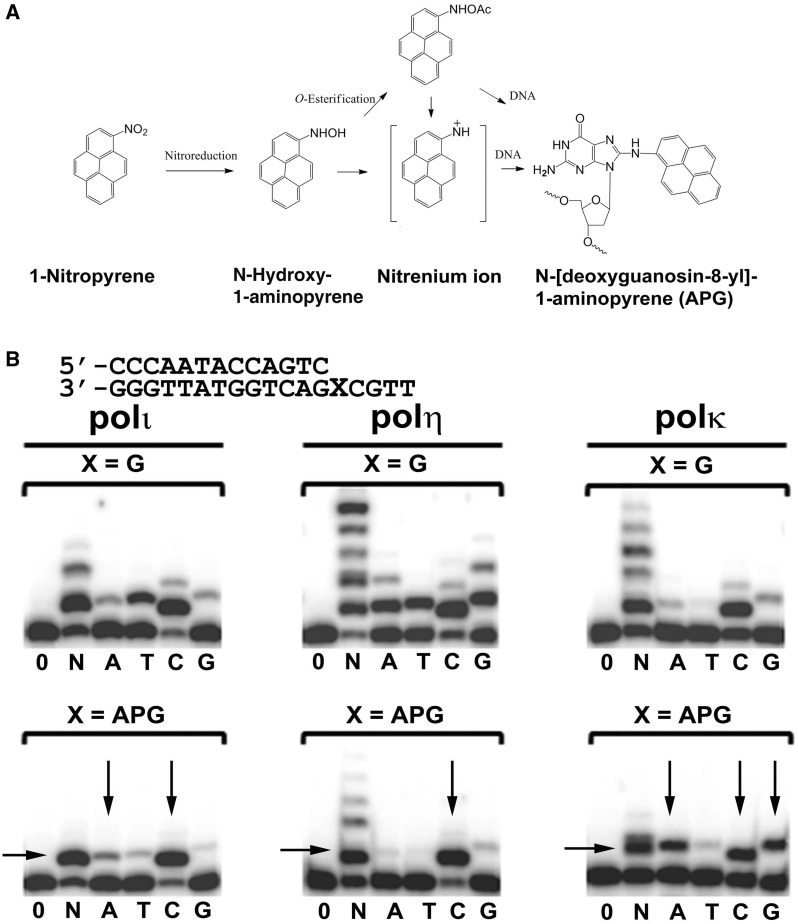
Urban air pollution increases morbidity and mortality rates in human populations (1). One of the main contributors to the detrimental health effects of air pollution is exposure to nitrated polycyclic aromatic hydrocarbons (NPAHs). NPAHs are a group of abundant organic chemical pollutants, arising from the combustion of carbon-containing agents, such as diesel exhaust, industrial emissions and cigarette smoke (2). The toxicity of NPAH compounds arises from their metabolic nitro-reduction in human cells, creating highly reactive species that react with genomic DNA (Figure 1A). 1-Nitropyrene (1-NP), the most prevalent NPAH in the environment, is particulalrly abundant in urban air particulate. 1-NP induces mutagenesis (3,4) and apoptosis (5) by forming DNA adducts in mammalian cells. 1-NP causes mammary gland tumors in experimental animals (6,7). Thus, 1-NP and related NPAH compounds are suspected to have a major impact on human health, especially in populations living in urban or industrial areas. Metabolites of 1-NP covalently bind to guanine bases in DNA, forming mainly the N-[deoxyguanosine-8-yl]-1-aminopyrene (APG) adduct (Figure 1A) (8,9). The mutagenic signature of the APG lesion is the induction of G to T transversions (3). G to T transversions are pronounced mutations in lung cancers from smokers, and high concentrations of NPAH compounds in cigarette smoke may be a contributing factor to the observed genetic changes (8,10).
Figure 1.
1-Nitropyrene and Y-family DNA polymerase activity. (A) Reduction of 1-Nitropyrene to the nitrenium ion and attachment to a guanine base of DNA to form APG adducts. (B) Primer extension assays with undamaged G or the APG lesion at the first replication position. Human polη, polκ and polι were incubated with DNA substrates and reacted with no incoming nucleotides (0), all four nucleotides (N) or individual nucleotides (A, T, C, G) for either 0.5 min for undamaged G or 30 min for the APG lesion. Vertical arrows indicate nucleotide preferences opposite the APG lesion, and horizontal arrows indicate stalling bands of APG DNA. DNA substrates are shown above the gels.
Bulky DNA lesions, such as APG, hinder DNA replication carried out by high-fidelity polymerases owing to the restrictive active site of these enzymes (11). To rescue adduct-stalled replication forks, cells must recruit specialized Y-family DNA polymerases, which replicate through bulky DNA lesions by translesion synthesis (12–14). Although these polymerases alleviate stalled replication forks, they are also highly error-prone and induce mutations. Y-family DNA polymerases have a finger, thumb and palm domain similar to all DNA polymerases, and a unique fourth domain referred to as the little finger (or polymerase-associated domain (PAD)/wrist) domain (15–19). The first three domains pack tightly togther and form a catalytic ‘core’ with a loose connection to the little finger domain (20,21). Y-family polymerases have open and solvent-exposed active sites, which can accommodate distorted and bulky DNA lesions, but are responsible for low-fidelity DNA replication (12,15). Multiple Y-family polymerases exist in most eukaryotic species, each with distinct functionalities (22). Human cells contain four Y-family members: Rev1, polymerase η (polη), polymerase ι (polι) and polymerase κ (polκ) (12,13). The last three are translesional DNA polymerases that differ in their ability to bypass lesions and their fidelity during DNA replication. Y-family DNA polymerases are thought to be responsible for the mutagenic signature of cells exposed to 1-NP.
To reveal the molecular basis of error-prone replication of the APG DNA adduct, we performed functional analysis on three human Y-family DNA polymerases in APG bypass, determined the structures of human polι in ternary complex with APG-containing DNA at the insertion stage and extended our structural observations of polι toward structural models for polη and polκ APG DNA adduct bypass in humans.
MATERIALS AND METHODS
Synthesis of oligonucleotides containing APG
The protected monomer 2,8-diisobutyryl-8 -(1-aminopyrenyl)-5′-O-(4,4′-dimethoxytrityl)-3′-O-[N,N′-diisopropylamino(2-cyanoethoxy)phosphonyl]-2′-deoxyguanosine was prepared as described (23). It was incorporated by standard DNA synthesis protocol into the oligonucleotide 5′-TCAG*GGGTCCTAGGACCC-3′ (where G* = APG). The mass of the 18-mer was confirmed by ESI-MS analysis.
Protein preparation
Polι protein used for crystallization was expressed and purified as previously described (24). Proteins used for replication assays (polι 1-430, polη 1-445, polκ 19-523, polη R61A 1-445) contained N-terminal histidine tags and were expressed in Escherichia coli and purified by nickel affinity, followed by ion-exchange, chromatography.
DNA preparation
The APG DNA substrates used for crystallization and activity assays were purified using ion-exchange chromatography (25). For polι crystallization, the self-annealing 18-mer oligonucleotide containing an APG (G) lesion (5′-TCAGGGGTCCTAGGACCC-3′) was annealed with itself to give a DNA substrate with two replicative ends. Undamaged oligonucleotides used for primer extension assays were purchased from Keck Oligo Inc. and purified by ion exchange. For primer extension assays, a 12-nt primer (5′-CCCAATACCAGTC-3′) was annealed to an 18-nt undamaged G or APG template (5′-TTGCGGACTGGTATTGGG-3′). For the extension assays, a 13-nt primer containing C at the 3′-end (5′-CCCAATACCAGTCC-3′) or A at the 3′-end (5′-CCCAATACCAGTCA-3′) was annealed to either the 18-nt undamaged template or the 18-nt APG template. Primers were 5′-end labeled using [γ-32P]ATP and T4 polynucleotide kinase and annealed to the template DNA substrates.
Primer extension assays
DNA substrates (10 nM) were incubated with either polι, polη, polκ or polη R61A (10 nM) and 100 uM of either all four dNTPs or individual dNTPs at 37°C in reaction buffer containing 40 mM Tris (pH 8.0), 5 mM MgCl2, 250 ug/ml bovine serum albumin, 10 mM DTT and 2.5% glycerol. For the primer extension assays, reactions were carried out for ∼2 min with undamaged DNA and ∼30 min for APG DNA. Reaction times for all other experiments were indicated below the gels. Reactions were terminated with loading buffer (95% formamide, 20 mM EDTA, 0.025% xylene, 0.025% bromophenol blue) and resolved on a 20% polyacrylamide gel containing 7 M urea. Gels were visualized using a PhosphorImager (Storm 860, GE Healthcare).
Crystallization and structure determination
Ternary complexes were formed for APG-dCTP and APG-dATP by incubating polι protein (0.2 mM) and DNA in a 1:1.2 ratio with dNTP (5 mM) and MgCl2 (5 mM). Crystals of both complexes were obtained in 15% PEG 5000 MME, 0.2 M (NH4)2SO4, 2.5% glycerol, 0.1 M MES (pH 6.5). Crystals were flash frozen in liquid nitrogen directly from dehydrated crystallization drops to prevent crystal cracking. X-ray diffraction data were collected at beamline 24-ID-E at the Advanced Photon Source in Argonne National Laboratory. All data were processed and scaled using HKL (26).
Both structures were solved by molecular replacement using PHASER (27), with a previously solved ternary complex (PDB: 3GV5) (iota) as a search model. Structural refinement was performed using PHENIX (28), starting with rigid-body refinement, followed by restrained postional and B-factor refinement, and lastly, TLS refinement (29). Model building was performed using COOT (30), and figures were created using PYMOL (31).
Modeling APG–polymerase complexes
For modeling the APG lesion:dNTP conformations in human DNA polη and human polκ, initial PDB structures of 3MR2 and 2OH2 were used for polη and polκ, respectively (17,18). Briefly, the polι:APG structures were superimposed with the structures of polη and polκ to install the APG substrate (DNA and dNTP) from polι into polη and polκ. The positions of the adducted guanine containing replicating base pairs were slightly adjusted in the active sites of the original polη and polκ structues. For modeling APG extension in polι, the APG-dCTP and APG-dATP structures were used as starting models. The substrates (DNA and dNTP) in the complex structure were translocated as a rigid body from the insertion position down to the extension position. Then, the undamaged DNA and replicating base pair from a polι structure (PDB: 2ALZ) were used as a reference to build up the replicating base pairs in the extension models.
RESULTS
Y-family polymerases bypass APG with different fidelities
To characterize the bypass capability and mutagenic potential of human Y-family DNA polymerases across the APG lesion, we carried out primer extension assays using human polι, polη and polκ with either undamaged G or the APG lesion in the template DNA strand. Opposite undamaged G, the three enzymes extend the primer with varying efficiencies, but each polymerase preferentially incorporates the correct C nucleotide (Figure 1B). Misincorporation bands are observed for all three enzymes, a trend noted with the Y-family DNA polymerases owing to their open and solvent-accessible active sites (15). In the presence of the APG lesion, all three enzymes display stalling of replication at the lesion site (strong stalling band indicated by horizontal arrows in Figure 1B), with polη showing the greatest ability to bypass the lesion (multiple bands above the stalling band in the presence of all four nucleotides). The strong stalling bands in primer extension assays indicate the low efficiency of the DNA polymerases to extend past the APG adduct after single nucleotide incorporation opposite the lesion. Recent kinetic studies have revealed that polη, polκ and polι have a 6-, 5.7- and 7-fold decrease, respectively, in replication efficiency opposite the APG lesion compared with undamaged G, with polη exhibiting the highest bypass efficiency, followed by polκ, which, in turn, was more efficient than polι (32). Although the efficiency of reaction is significantly reduced, all three DNA polymerases preferentially incorporate the correct C nucleotide opposite the APG lesion (Figure 1B). Polη and polι appear to have higher fidelities opposite the APG lesion relative to an undamaged G. In contrast, polκ has lower fidelity for the lesion, and significant A and G misincorporations opposite APG occurred (Figure 1B). Interestingly, the misincorporation of A by both polι and polκ increased opposite the APG lesion compared with undamaged G. Indeed, kinetic experiments have revealed a 1.4- and 11-fold increase in A incorporation opposite APG for polι and polκ, respectively. The increase in A misincorporation is striking considering that the mutagenic signature of the APG adduct is G to T transversions induced by A mismatches. These results indicate that Y-family DNA polymerases are likely to cause mutations during cellular replication of the APG lesion.
Polι-APG-dNTP ternary structures: conformational changes on APG binding
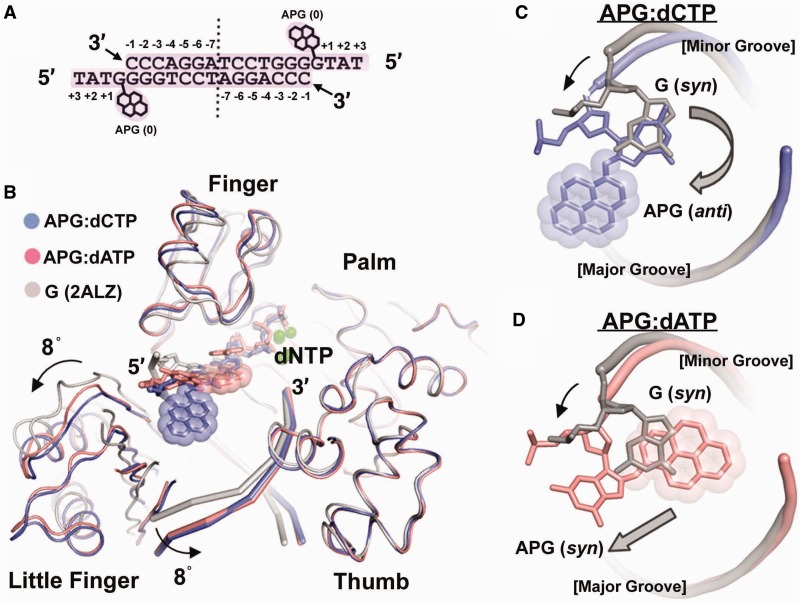
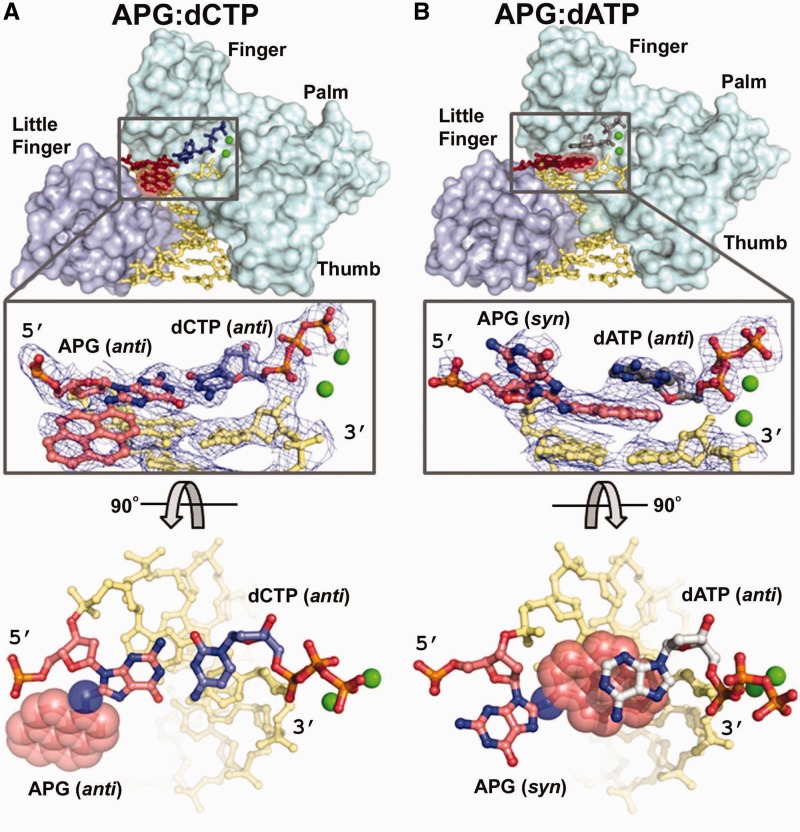
To elucidate the mechanism of replication stalling and A misincorporation opposite the APG lesion by polι, we crystallized polι in complex with APG DNA incorporating either dCTP or dATP nucleotides directly opposite the lesion. The DNA substrate for crystallization was designed so that the lesion was located directly downstream to the primer–template junction, ready for dNTP incorporation (Figure 2A). The DNA substrate was incubated with polι and co-crystallized with either incoming dCTP or dATP nucleotide. The resulting structures are denoted as APG-dCTP and APG-dATP, according to the identity of the incoming nucleotides in the active site. Both polι-APG ternary crystals diffracted to 2.9 Å resolution (Table 1), which represent the first set of structures of a DNA polymerase replicating directly opposite a bulky pyrene lesion at the insertion stage. The polι-APG structures have the same crystal form as previously solved polι structures with undamged G (33); the asymmetric unit contains one polι and one-half of the DNA duplex from a self-annealed DNA oligonucleotide (see ‘Materials and Methods’ section). The complexes are both in productive conformations, with the α phosphate of dCTP/dATP in reach of the 3′-OH of the primer strand (Figures 2 and 3). Electron density is observed in the APG-dCTP structure linking the 3′-OH of the primer strand to the α phosphate, suggesting that some reaction intermediates or products have been generated in the crystal (Figure 3A). The 3′-OH group of the primer strand is 2.9 Å from the α phosphate in APG-dCTP. However, in the APG-dATP structure, the majority of the complexes appear to be unreacted, with disconnected electron density between the 3′-end of the primer and the α phosphate, implying that the polymerase bypass of the APG lesion is difficult and inefficient (Figure 3B). The 3′-OH group of the primer strand is ∼3.5 Å away from the α-phosphate oxygen, a result of an unusual dATP base positioning (details in the next section), which likely reduces the efficiency of reaction. For both APG structures, two magnesium (Mg) ions are observed in the active site at similar positions to an undamaged DNA polι ternary structure (2ALZ). Briefly, in the APG-dCTP structure, the B-position Mg ion is coordinated by the β (3.0 Å)- and γ (2.5 Å)-phosphate oxygen atoms, whereas the A position Mg ion is coordinated by the α (2.8 Å)-phosphate oxygen and the primer strand’s 3′-OH (3.2 Å), as well as surrounding O atoms from polι and solvent (detailed bonding distance listed in Supplementary Figure S1A). For APG-dATP, the B-position Mg ion is coordinated by the β (2.7 Å)- and γ (2.4 Å)-phosphate oxygen atoms, whereas the A-position Mg ion is coordinated by the α (2.4 Å)-phosphate oxygen and the 3′-OH group of the primer strand (3.2 Å), along with other oxygen atoms from polι and solvent (Supplementary Figure S1B).
Figure 2.
Comparisons of polι-APG ternary structures. (A) DNA substrate used for crystallization and the positions of bases. Numbering is relative to the APG template at position 0. Vertical dashed line indicates axis of 2-fold symmetry. (B) Superposition of APG-dCTP (blue), APG-dATP (pink) and a previous polι ternary complex with undamaged G (PDB: 2ALZ, grey). Domains are labeled and arrows indicate domain movement relative to the undamaged G structure. The aminopyrene lesion is shown with incoming nucleotides and metal ions (green spheres). (C) Positioning of APG in APG-dCTP (blue) and (D) APG-dATP (pink) relative to undamged G (grey). View is looking from top, down through the DNA helix. Black arrows indicate backbone DNA movement, and grey block arrows indicate APG base movement. Major and minor groove sides of the DNA helices are labeled.
Table 1.
Summary of crystallographic data
| Data collection | APG-dCTP | APG-dATP |
|---|---|---|
| Space group | P6522 | P6522 |
| Mol/AUa | 1 | 1 |
| Unit cell | ||
| a, b, c (Å) | 98.0, 98.0, 194.6 | 98.9, 98.9, 194.2 |
| α, β,γ (°) | 90, 90, 120 | 90, 90, 120 |
| Resolution (Å)b | 32.0–2.90 (2.95–2.90) | 32.1–2.90 (2.95–2.90) |
| Unique reflections | 13 253 | 13 665 |
| Completeness (%)b | 97.2 (98.2) | 98.5 (98.8) |
| Redundancyb | 7.0 (7.2) | 4.0 (4.2) |
| I/σIb | 29.4 (2.7) | 27.5 (2.7) |
| Rmergeb | 7.8 (63.3) | 6.2 (55.2) |
| Refinement statistics | ||
| Rwork/Rfree | 23.9/28.1 | 21.8/26.4 |
| Number of atoms | ||
| Protein | 2853 | 2951 |
| DNA | 326 | 326 |
| dNTP | 29 | 30 |
| Ionsc | 3 | 3 |
| Water | 28 | 28 |
| Average B factor | ||
| Protein | 85.6 | 89.7 |
| DNA | 88.8 | 79.9 |
| dNTP | 111.4 | 106.2 |
| Ions | 81.5 | 82.8 |
| Waters | 62.2 | 88.8 |
| R.m.s. deviations | ||
| Bonds (Å) | 0.007 | 0.006 |
| Angles (°) | 1.17 | 1.14 |
aMol/AU represents the number of molecules per asymmetric unit.
bData in the highest resolution shell are in parentheses.
cOne non-catalytic Mg2+ ion exists in both structures.
Figure 3.
The polι-APG ternary complexes with zoomed-in replicating base pairs (A) APG-dCTP and (B) APG-dATP structures. The finger, thumb and palm domains are colored cyan as the ‘core’, while the little finger domains are colored light blue. The DNA is in yellow, the APG lesion in red, dCTP in blue and dATP in grey. Active site metal ions are shown as green spheres. Zoom-in views of the active sites are shown below the structures with the 2Fo-Fc electron density map contoured at 1σ and in top views of the APG:dCTP and APG:dATP replicating base pairs. The APG lesion is in red, with sphere representation for the hydrophobic ring in the top views.
The overall structures of two APG complexes look similar to each other and previous undamaged DNA-polι structures (Figure 2B). However, small differences are noted in the finger positioning, resulting from conformational differences in APG and the incoming nucleotides in the active site. The front of the finger domain of APG-dATP is pushed up by ∼2 Å to accommodate dATP that is off the regular position of an incoming nucleotide (Figure 2B). It is noteworthy that significant conformational changes are observed in the little finger domains and DNA substrates in both APG complexes (Figure 2B), comparing the APG polι structures with a previously solved polι ternary complex with undamaged G (2ALZ) (33). The little finger domains have moved downward by ∼8° relative to the undamaged G structure, in response to the 5′-end of the template DNA moving toward the solvent-exposed major groove (Figure 2B). Consequently, the bottom of the DNA substrate has rotated toward the thumb domain by ∼8° to accommodate the shift in the little finger. The polι-APG structures share the same crystal form as previously solved polι structures (2ALZ) with undamged G, with the largest interface in the crystal between the protein and DNA substrate. These observations suggest that the domain and DNA movements are controlled by the complex structure, not the packing environment of the crystal lattice. The domain and DNA re-orientations in our APG structures are the result of the adjustments necessary for the bulky APG lesion to be accommodated within the polι active site. Such adjustments of the substrate and little finger domain positions have been previously observed with Dpo4, the model DNA polymerase in the Y family, as well as with yeast polη and polκ structures (18,20,21,34). This flexible adjustment of the little finger domain is a common structural characteristic of the Y-family DNA polymerases, as the little finger has loose connections to the rest of the polymerase core (20,21). The finger domain movement in APG-dATP, however, is not a common structural observaton. The active sites of Y-family polymerases, mainly defined by the finger domains, have been observed in a pre-formed and rigid state (15,20). The finger of Dpo4 does not open up even when replicating double base lesions, such as CPD (TT dimer) and cisplatin-linked Pt-GG, which forces the two cross-linked bases to squeeze into the active site (35,36). However, our current polι structures are the first to show a productive bulky adduct DNA in the active site of a Y-family polymerase and possibly reveal a new structural plasticity within this polymerase family.
APG adduct and incoming nucleotide: dNTP-induced APG conformational changes
The conformation of the adducted G was dramatically altered compared with undamaged G in the polι active site (33) (Figure 2). Moreover, the conformation of the APG lesion was vastly different in the two APG complexes, which appears to be directly influenced by the identity of the incoming nucleotides. Undamaged template purines adopt syn conformations to form a Hoogsteen base pair with incoming nucleotides, orienting the C8 atom toward the protein-occluded minor groove in the polι active site (33) (Figure 2). Polι induces syn conformations owing to a remarkably narrow active site that restricts the C1′–C1′ distance to <9 Å. A Watson–Crick base pair requires a C1′–C1′ distance of ∼10.6 Å, and thus, this mode of base pairing is highly unfavorable in the narrow polι active site (24,33). Because the bulky APG ring is linked through the C8 atom of guanine, a syn conformation would result in a clash of the bulky pyrene ring into the protein-occluded minor groove side if the modified G remains in the active site. To avoid the steric conflicts, the APG base in the APG-dCTP structure adopts an anti conformation different from a regular G template, with the APG ring placed in the solvent-exposed major groove (designated as an extra-helical conformation, Figures 2C and 3A). The APG base and DNA backbone are shifted out toward the major groove to achieve a standard C1′–C1′ distance for anti-guanine:dCTP Watson–Crick base pairing (Figure 2C and 3A). This observation is consistent with the prediction that Watson–Crick base pairing could occur in the polι active site for a major groove adduct by a previous modeling study (37). Thus, our APG-dCTP structure illustrates how polι can accommodate the APG lesion in Watson–Crick base pairing, a mechanism different from Hoogsteen base pairing observed in all the previous purine template structures of polι (33). In this orientation, the hydrophobic aminopyrene moiety is positioned in the solvated major groove, with no direct interactions with either polι or the DNA substrate, generating a mobile pyrene ring with high B factors (∼120Å2) compared with the rest the protein/DNA (∼88Å2). Previous solution NMR experiments have revealed that the APG lesion opposite C is favored in an intra-helical conformation within the DNA helix in the absence of a protein (38). Thus, polι must force APG into an energetically unfavorable conformation to create a productive complex with dCTP, which likely contributes to the low reaction efficiency.
In the APG-dATP structure, however, the APG-adducted G base maintains a syn conformation but projects itself into the major groove, allowing the bulky pyrene ring to be placed in the replicating base pair position within the DNA helix (designated as an intra-helical conformation, Figures 2D and 3B). The bulky attachment in APG is sandwiched between the underlying base pair and the incoming dATP (Figure 3B). Interestingly, the NMR structure of a DNA helix containing the APG lesion opposite dA revealed an almost identical APG conformation with a syn glycosidic bond and an intercalated pyrene ring (39). Thus, intra-helical APG conformations represent a low-energy state of the lesion that stabilizes the hydrophobic pyrene ring. Previously observed intercalated pyrene rings in complex with Dpo4 polymerase reveal primer strands and incoming dNTPs that are separated by up to 10 Å and generate non-productive conformations for lesion bypass (40,41). The APG-dATP structure in this work provides the first intercalated pyrene ring in a productive complex with a Y-family polymerase. The intercalated pyrene ring occupies the space of the replicating base pair and prevents the incoming dATP from entering the active site in plane with the adducted template (Figure 3B). Consequently, the base of dATP is forced up one base pair position and stacks on top of the pyrene ring, while its phosphate moiety maintains a regular position as in other polι structures (Figures 2B and 3B) (24,33). No hydrogen bonds are formed between the dATP base and any template bases. Thus, dATP misincorporation is dictated purely by base-stacking interactions with the intercalated APG bulky ring (Figure 3B). The dATP base has the greatest stacking potential of all four nucleotides and thus would be most favored for stacking above pyrene rings (42). This observation is analogous to preferential incorporation of dATP on blunt-end DNA through stacking interactions by a Y-family polymerase (43). Accordingly, the intercalated APG ring mimics a base pair in the blunt end of a DNA helix and promotes A misincorporations. This observation also indicates how the APG lesion can induce frameshift mutations within the genome, a common occurrence with this type of DNA damage (44). Because the dATP stacks above the APG lesion, it is likely that base pairing could occur with template nucleotides above the APG base leading to −1 or −2 frameshift mutations. Thus, the strong stacking potential of the A base provides mechanistic insight into abundant G to T transversions and frameshift mutations (10).
Structural basis of APG bypass fidelities in different Y-family polymerases
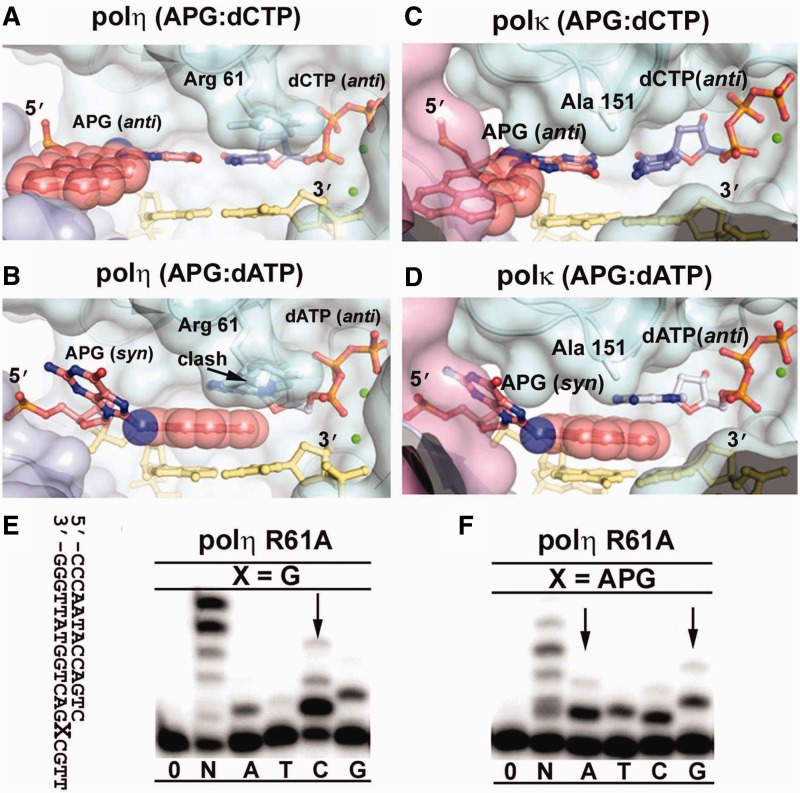
To understand differences in APG incorporation specificity between different human Y-family polymerases, we modeled the polι APG base pairs into the active sites of polη and polκ. Modeling indicates that polη would be able to accommodate dCTP with APG in the extra-helical conformation (Figure 4A), but not the stacked dATP with APG in an intra-helical conformation (Figure 4B). The Arg 61 residue located on the lid of the finger domain makes the active site of polη more restrictive than polι and would clash with the dATP base stacking over the pyrene ring (Figure 4B), preventing A base misinsertion. The residues on the lid of the finger domain contact the replicating base pair in the active site and control the substrate specificity of Y-family polymerases (15). The model provides structural insight into the low A misinsertion frequency of polη opposite APG compared with the other human polymerases in our replication assays (Figure 1B). To validate this structural model, Arg 61 of polη was replaced with Ala by site-specific mutagenesis. This mutation causes a loss of fidelity opposite APG, with greatly enhanced A misincorporations (Figure 4F), but it does not reduce the fidelity opposite undamaged G (Figure 4E). Thus, the unique active site of polη prevents A misincorporations opposite APG by inhibiting productive incoming nucleotide complexes with the intra-helical APG conformation.
Figure 4.
Modeling of polη and polκ APG replication. The APG:dCTP Watson–Crick base pair (A and C) and the APG:dATP stacking base pair (B and D) from DNA in APG-dCTP and APG-dATP were modeled into polη (3MR2) and polκ(2OH2), respectively. The color scheme is identical to Figure 3. The pink domain in (B and D) is the N-clasp of polκ, which shadows the little finger domain, but does not directly contact DNA. Arg 61 of polη clashing with dATP and Ala 151 of polκ close to dATP are labeled. Replication fidelity of the polη R61A mutant for undamaged G (E) and the APG lesion (F). Polη R61A was incubated with DNA substrates and reacted with no incoming nucleotides (0), all four nucleotides (N) or individual nucleotides (A, T, C, G) for 1 min. The DNA substrate is shown left to the gels. Arrows indicate incoming nucleotide preference to G and increased misinsertion opposite APG.
Modeling the APG lesion in polκ reveals that a mismatched A base would be able to stack above the intercalated APG lesion similar to polι owing to small residues (Ala 150 and Ala 151) in the polκ’s finger domain lid, which contacts the replicating base pair (Figure 4C and D). A slight shift up of the finger domain, similar to polι (Figure 2B), would enable polκ to accommodate the intra-helical APG and stacked dATP in its active site (Figure 4D). In addition, polκ has an additional N-clasp to cover the major groove near the active site that is fully exposed to the solvent in other Y-family polymerases. The N-clasp coverage may also contribute to the bulky lesion bypass by protecting the hydrophobic bulky lesion and bigger purine bases of incoming nucleotides from aqueous solvent. The model could explain the low fidelity of APG replication by polκ observed in the function assays, particularly for misinsertion of purine bases A and G with high stacking potentials against APG (Figure 1B). Nevertheless, the finger domains of Y-family polymerases play an important role in replication fidelity and lesion bypass specificity, as the finger domains directly contact the replicating base pair in the active sites (15,24). Indeed, swapping finger domains between polι and Dpo4 Y-family polymerases results in exchanges of the fidelity and specificity of the enzymes (24). The sequence of the finger domain varies among Y-family polymerases, which generates unique active sites with different shapes, charge distributions and flexibilities, allowing for distinctive specificity and activity during DNA replication (24).
APG:dC base pairing induces replication stalling
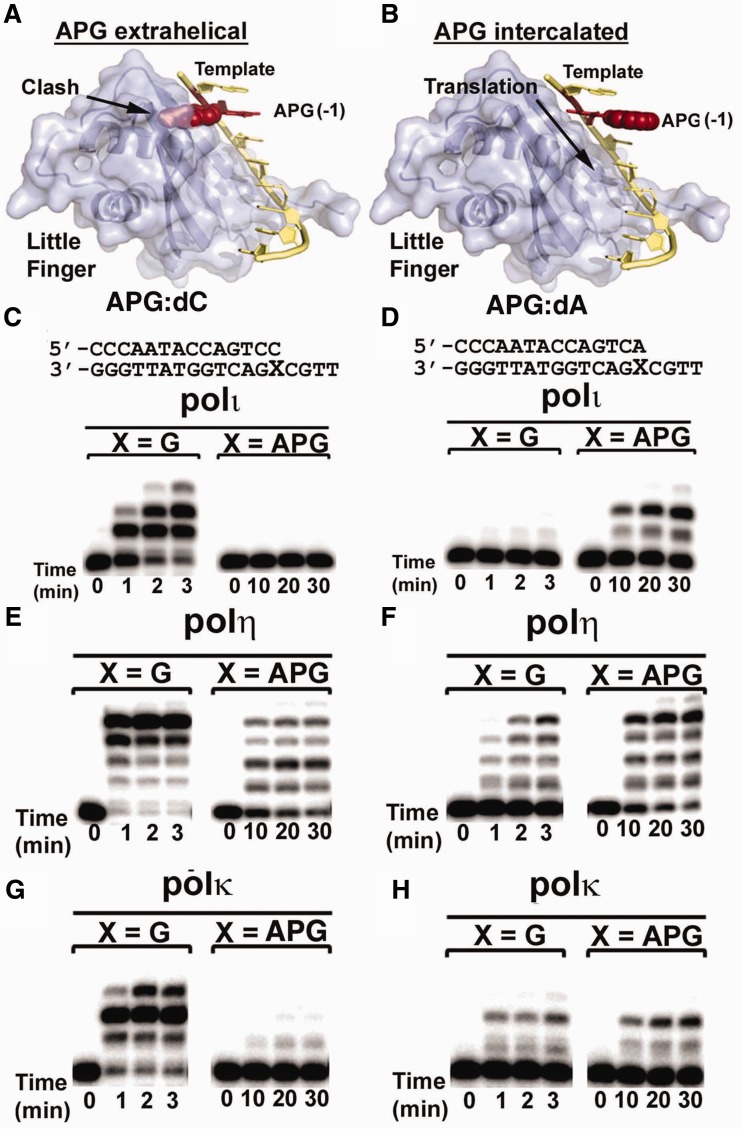
To understand how human Y-family polymerases would elongate a primer strand after the APG lesion, we performed replication assays to extend the primer strand beyond APG paired with correct C or mismatched A at the primer–template junction. Polι efficiently extended the primer strand from a regular G:C base pair, but was unable to extend from a lesion APG:C base pair (Figure 5C). Recent kinetic experiments have revealed that polι has ∼4500-fold decrease in extension efficiency past the APG:C lesion base pair compared with undamaged G (32). Interestingly, when an A nucleotide was mispaired with G/APG bases, a complete opposite extension effect was observed: primer extension was inhibited by a G:A mismatch, whereas a lesion APG:A mismatch was efficiently extended by polι (Figure 5D). For APG lesion DNA replication, a similar trend of the APG:C stalling and APG:A extension was observed for polκ (Figure 5G and H) and polη, albeit to a lesser extent for polη (Figure 5E and F). The kinetic studies have shown an 18- and 200-fold reduction in APG:C extension efficiency for polη and polκ, respectively, indicating that the difficulty in extending the APG lesion paired to the correct C nucleotide exists for all polymerases, although the magnitude varies for different polymerases. To understand the differences in APG extension, we modeled APG:C and APG:A base pairs observed in our structures into the extension position (-1 position) in polι. At the -1 position, the extra-helical APG lesion from APG-dCTP clashes with the little finger domain (Figure 5A). This structural conflict would inhibit the translocation of the template DNA through the polymerase and block primer elongation. In contrast, the intercalated APG lesion from the APG-dATP structure can freely translocate into the -1 position, with no inhibitory protein interactions, because the APG has no structural conflicts with polι (Figure 5B). These results suggest that the incoming nucleotide-induced APG conformation plays a critical role in primer extension after the APG lesion.
Figure 5.
Effects of APG conformations on primer extension beyond APG lesion. Modeling of template DNA translocation through polι with extra-helical (A) and intra-helical (B) APG conformations. Template DNA is shown in yellow, the APG lesion at the -1 extension position is shown in red and the little finger domain is shown in light blue. Polι, polη and polκ were incubated with undamaged G or the APG lesion paired with correct C (C, E, G) or mismatched A (D, F, H) from the primer strand. Reactions were carried out with all four nucleotides at various time points, as indicated under each lane. DNA substrates are shown above the gels.
DISCUSSION
NPAHs are highly abundant environmental pollutants with detrimental effects on human health. The structures presented here provide the first indication of how a human DNA polymerase may replicate directly opposite an NPAH-derived DNA adduct. The structures also reveal the mechanism of how a major NPAH–guanine lesion can induce A misincorporations. The hydrophobic ring systems of APG, and presumably other PAH/NPAH lesions, have the ability to stack above the primer–template junction in an intra-helical conformation to achieve a thermodynamically stable structure. This conformation, in turn, allows the mismatched A nucleotide to stack above the ring system and be incorporated into the growing primer strand in a template-independent manner. After one more round of DNA replication, the A-mismatched base pair will result in the signature G to T transversion. Although the size of the NPAH ring system and different chemical attachments could alter the base-stacking potential in the DNA helix, the stacking mechanism of A misincorporation is likely to be common to all genotoxic PAH/NPAH DNA lesions. We speculate that a common mechanism of A misincorporation may explain the widespread mutagenic potential of PAH/NPAH compounds, and their propensity to elicit carcinogenesis.
Interestingly, A misincorporations opposite NPAH–guanine lesions may provide cells with a survival advantage over correct C incorporation under certain pathological conditions. Our structural and biochemical studies have revealed that the mismatched A nucleotide promotes primer elongation by stabilizing the NPAH ring system in the DNA helix, whereas the correctly matched C nucleotide induces replication stalling by projecting the NPAH lesion into the protein molecule by polι and polκ. Thus, correct C incorporation may induce replication fork stalling under certain situations, such as with non-functional polη. The stalling may contribute to the cellular toxicity observed for NPAH compounds (5). In contrast, the mismatched A nucleotide would likely promote replication fork progression by polι or polκ, allowing cell survival but leading to high rate of mutagenesis (10). This type of error-prone lesion bypass is a hallmark of Y-family polymerase function and is an evolutionarily conserved mechanism that allows replication fork progession during times of cellular stress.
One question that remains to be answered is which Y-family polymerase is the predominant enzyme for NPAH DNA lesion bypass. It is likely that multiple DNA polymerases, particularly translesional Y-family polymerases, participate in NPAH lesion bypass in mammalian cells (45). A two-polymerase model has been proposed for DNA lesion bypass, in which one polymerase performs insertion and another performs primer extension for each lesion bypass event (46). From our biochemical experiments and previous work by other groups, it appears that polη is the predominant polymerase for ‘error-free’ APG lesion bypass. However, it is important to note that many cancers associated with exposure to air-borne carcinogens, particularly lung and esophageal cancers, are either linked to the polι gene or have significant polι over-expression (47–49). Therefore, in pathological situations where the proportions of Y-family polymerases are dysregulated, it is likely that an over-abundance of polι causes a higher frequency of NPAH lesions to be bypassed by error-prone polι rather than relatively error-free polη, leading to increased genetic mutations. Indeed, it has been shown that the high rates of mutations observed in xeroderma pigmentosum variant syndrome are due to polι taking over the bypass roles of polη (50). Furthermore, mutational burdern in breast cancer cell lines (which has been linked to aminopyrene exposure in animal models) is directly correlated to polι expression (51). Our structural observations combined with these lesion-induced mutagenesis results suggest that polι could play an important role in 1-NP–derived mutations in human cells.
Although there are different carcinogenic NPAH and PAH compounds found in polluted air, the metabolites of these compounds all share three common characteristics of hydrophobicity, preferential attachment to guanine nucleotides and induction of G to T transversions at the site of the guanine adduct (52–56). Thus, the results presented herein would likely be applicable to a wide variety of bulky DNA lesions and provide mechanistic insight into the health dangers imposed by this type of chemical air pollution.
ACCESSION NUMBERS
The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org, with accession codes 4EYH and 4EYI for APG-dCTP and APG-dATP, respectively.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figure 1.
FUNDING
Canadian Institutes of Health Research (CIHR) operating grant [MOP93590 to H.L.]; National Institute of Environmental Health Sciences grant [ES09127 to A.K.B.]. Funding for open access charge: CIHR.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank beamline support at 24-ID of APS in Argonne National Laboratory.
REFERENCES
- 1.Samet JM, Dominici F, Curriero FC, Coursac I, Zeger SL. Fine particulate air pollution and mortality in 20 U.S. cities, 1987-1994. N. Engl. J. Med. 2000;343:1742–1749. doi: 10.1056/NEJM200012143432401. [DOI] [PubMed] [Google Scholar]
- 2.Scheepers PTJ, Velders DD, Martens MHJ, Noordhoek J, Bos RP. Gas-chromatographic mass-spectrometric determination of nitro polycyclic aromatic-hydrocarbons in airborne particulate matter from workplace atmospheres contaminated with diesel exhaust. J. Chromatogr. A. 1994;677:107–121. [Google Scholar]
- 3.Watt DL, Utzat CD, Hilario P, Basu AK. Mutagenicity of the 1-nitropyrene-DNA adduct N-(deoxyguanosin-8-yl)-1-aminopyrene in mammalian cells. Chem. Res. Toxicol. 2007;20:1658–1664. doi: 10.1021/tx700131e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andersson H, Piras E, Demma J, Hellman B, Brittebo E. Low levels of the air pollutant 1-nitropyrene induce DNA damage, increased levels of reactive oxygen species and endoplasmic reticulum stress in human endothelial cells. Toxicology. 2009;262:57–64. doi: 10.1016/j.tox.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 5.Asare N, Landvik N, Lagadic-Gossmann D, Rissel M, Tekpli X, Ask K, Lag M, Holme J. 1-Nitropyrene (1-NP) induces apoptosis and apparently a non-apoptotic programmed cell death (paraptosis) in Hepa1c1c7 cells. Toxicol Applied Pharmacol. 2008;230:175–186. doi: 10.1016/j.taap.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 6.Hirose M, Lee MS, Wang CY, King CM. Induction of rat mammary-gland tumors by 1-Nitropyrene, a recently recognized environmental mutagen.GEN. Cancer Res. 1984;44:1158–1162. [PubMed] [Google Scholar]
- 7.Elbayoumy K, Rivenson A, Johnson B, Dibello J, Little P, Hecht SS. A study of chemical carcinogenesis .114. comparative tumorigenicity of 1-nitropyrene, 1-nitrosopyrene, and 1-Aminopyrene administered by gavage to Sprague-Dawley rats. Cancer Res. 1988;48:4256–4260. [PubMed] [Google Scholar]
- 8.Djuric Z, Fifer E, Yamazoe Y, Beland F. DNA-binding by 1-nitropyrene and 1,6-dinitropyrene invitro and invivo - effects of nitroreductase induction. Carcinogenesis. 1988;9:357–364. doi: 10.1093/carcin/9.3.357. [DOI] [PubMed] [Google Scholar]
- 9.Sherrer S, Brown J, Pack L, Jasti V, Fowler J, Basu A, Suo Z. Mechanistic studies of the bypass of a bulky single-base lesion catalyzed by a Y-family DNA polymerase. J. Biol. Chem. 2009;284:6379–6388. doi: 10.1074/jbc.M808161200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hainaut P, Pfeifer GP. Patterns of p53 G–>T transversions in lung cancers reflect the primary mutagenic signature of DNA-damage by tobacco smoke. Carcinogenesis. 2001;22:367–374. doi: 10.1093/carcin/22.3.367. [DOI] [PubMed] [Google Scholar]
- 11.Hsu GW, Huang X, Luneva NP, Geacintov NE, Beese LS. Structure of a high fidelity DNA polymerase bound to a benzo[a]pyrene adduct that blocks replication. J. Biol. Chem. 2005;280:3764–3770. doi: 10.1074/jbc.M411276200. [DOI] [PubMed] [Google Scholar]
- 12.Yang W, Woodgate R. What a difference a decade makes: insights into translesion DNA synthesis. Proc. Natl Acad. Sci. USA. 2007;104:15591–15598. doi: 10.1073/pnas.0704219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedberg EC, Wagner R, Radman M. Specialized DNA polymerases, cellular survival, and the genesis of mutations. Science. 2002;296:1627–1630. doi: 10.1126/science.1070236. [DOI] [PubMed] [Google Scholar]
- 14.Sale JE, Lehmann AR, Woodgate R. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat. Rev. Mol. Cell Biol. 2012;13:141–152. doi: 10.1038/nrm3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ling H, Boudsocq F, Woodgate R, Yang W. Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell. 2001;107:91–102. doi: 10.1016/s0092-8674(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 16.Nair DT, Johnson RE, Prakash S, Prakash L, Aggarwal AK. Replication by human DNA polymerase-iota occurs by Hoogsteen base-pairing. Nature. 2004;430:377–380. doi: 10.1038/nature02692. [DOI] [PubMed] [Google Scholar]
- 17.Biertumpfel C, Zhao Y, Kondo Y, Ramon-Maiques S, Gregory M, Lee JY, Masutani C, Lehmann AR, Hanaoka F, Yang W. Structure and mechanism of human DNA polymerase eta. Nature. 2010;465:1044–1048. doi: 10.1038/nature09196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lone S, Townson SA, Uljon SN, Johnson RE, Brahma A, Nair DT, Prakash S, Prakash L, Aggarwal AK. Human DNA polymerase kappa encircles DNA: implications for mismatch extension and lesion bypass. Mol. Cell. 2007;25:601–614. doi: 10.1016/j.molcel.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 19.Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Rev1 employs a novel mechanism of DNA synthesis using a protein template. Science. 2005;309:2219–2222. doi: 10.1126/science.1116336. [DOI] [PubMed] [Google Scholar]
- 20.Wong JH, Fiala KA, Suo Z, Ling H. Snapshots of a Y-family DNA polymerase in replication: substrate-induced conformational transitions and implications for fidelity of Dpo4. J. Mol. Biol. 2008;379:317–330. doi: 10.1016/j.jmb.2008.03.038. [DOI] [PubMed] [Google Scholar]
- 21.Xing G, Kirouac K, Shin YJ, Bell SD, Ling H. Structural insight into recruitment of translesion DNA polymerase Dpo4 to sliding clamp PCNA. Mol. Microbiol. 2009;71:678–691. doi: 10.1111/j.1365-2958.2008.06553.x. [DOI] [PubMed] [Google Scholar]
- 22.Friedberg EC. Why do cells have multiple error-prone DNA polymerases? Environ. Mol. Mutagen. 2001;38:105–110. doi: 10.1002/em.1059. [DOI] [PubMed] [Google Scholar]
- 23.Colis LC, Chakraborti D, Hilario P, McCarty C, Basu AK. Synthesis of oligonucleotides containing 2′-deoxyguanosine adducts of nitropyrenes. Nucleosides Nucleotides Nucleic Acids. 2009;28:67–77. doi: 10.1080/15257770902736426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirouac KN, Ling H. Structural basis of error-prone replication and stalling at a thymine base by human DNA polymerase iota. EMBO J. 2009;28:1644–1654. doi: 10.1038/emboj.2009.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shanagar J. Purification of a synthetic oligonucleotide by anion exchange chromatography: method optimisation and scale-up. J. Biochem. Biophys. Methods. 2005;64:216–225. doi: 10.1016/j.jbbm.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 26.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 27.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr. D. Biol. Crystallogr. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 28.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Painter J, Merritt E. TLSMD web server for the generation of multi-group TLS models. J. Chromatogr. 2006;39:109–111. [Google Scholar]
- 30.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 31.DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA, USA: DeLano Scientific; 2002. [Google Scholar]
- 32.Sherrer SM, Sanman LE, Xia CX, Bolin ER, Malik CK, Efthimiopoulos G, Basu AK, Suo Z. Kinetic analysis of the bypass of a bulky DNA lesion catalyzed by human Y-family DNA polymerases. Chem. Res. Toxicol. 2012;25:730–740. doi: 10.1021/tx200531y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Human DNA polymerase iota incorporates dCTP opposite template G via a G.C + Hoogsteen base pair. Structure. 2005;13:1569–1577. doi: 10.1016/j.str.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 34.Silverstein TD, Johnson RE, Jain R, Prakash L, Prakash S, Aggarwal AK. Structural basis for the suppression of skin cancers by DNA polymerase eta. Nature. 2010;465:1039–1043. doi: 10.1038/nature09104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wong JH, Brown JA, Suo Z, Blum P, Nohmi T, Ling H. Structural insight into dynamic bypass of the major cisplatin-DNA adduct by Y-family polymerase Dpo4. EMBO J. 2010;29:2059–2069. doi: 10.1038/emboj.2010.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ling H, Boudsocq F, Plosky BS, Woodgate R, Yang W. Replication of a cys–syn thymine dimer at atomic resolution. Nature. 2003;424:1083–1087. doi: 10.1038/nature01919. [DOI] [PubMed] [Google Scholar]
- 37.Donny-Clark K, Shapiro R, Broyde S. Accommodation of an N-(deoxyguanosin-8-yl)-2-acetylaminofluorene adduct in the active site of human DNA polymerase iota: Hoogsteen or Watson-Crick base pairing? Biochemistry. 2009;48:7–18. doi: 10.1021/bi801283d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mao B, Vyas RR, Hingerty BE, Broyde S, Basu AK, Patel DJ. Solution conformation of the N-(deoxyguanosin-8-yl)-1-aminopyrene ([AP]dG) adduct opposite dC in a DNA duplex. Biochemistry. 1996;35:12659–12670. doi: 10.1021/bi961078o. [DOI] [PubMed] [Google Scholar]
- 39.Gu Z, Gorin A, Krishnasamy R, Hingerty BE, Basu AK, Broyde S, Patel DJ. Solution structure of the N-(deoxyguanosin-8-yl)-1-aminopyrene ([AP]dG) adduct opposite dA in a DNA duplex. Biochemistry. 1999;38:10843–10854. doi: 10.1021/bi9912138. [DOI] [PubMed] [Google Scholar]
- 40.Bauer J, Xing G, Yagi H, Sayer JM, Jerina DM, Ling H. A structural gap in Dpo4 supports mutagenic bypass of a major benzo[a]pyrene dG adduct in DNA through template misalignment. Proc. Natl Acad. Sci. USA. 2007;104:14905–14910. doi: 10.1073/pnas.0700717104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ling H, Sayer JM, Plosky BS, Yagi H, Boudsocq F, Woodgate R, Jerina DM, Yang W. Crystal structure of a benzo[a]pyrene diol epoxide adduct in a ternary complex with a DNA polymerase. Proc. Natl Acad. Sci. USA. 2004;101:2265–2269. doi: 10.1073/pnas.0308332100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guckian KM, Schweitzer BA, Ren RX, Sheils CJ, Tahmassebi DC, Kool ET. Factors contributing to aromatic stacking in water: evaluation in the context of DNA. J. Am. Chem. Soc. 2000;122:2213–2222. doi: 10.1021/ja9934854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fiala KA, Brown JA, Ling H, Kshetry AK, Zhang J, Taylor JS, Yang W, Suo Z. Mechanism of template-independent nucleotide incorporation catalyzed by a template-dependent DNA polymerase. J. Mol. Biol. 2007;365:590–602. doi: 10.1016/j.jmb.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Malia SA, Vyas RR, Basu AK. Site-specific frame-shift mutagenesis by the 1-nitropyrene-DNA adduct N-(deoxyguanosin-8-y1)-1-aminopyrene located in the (CG)3 sequence: effects of SOS, proofreading, and mismatch repair. Biochemistry. 1996;35:4568–4577. doi: 10.1021/bi9525132. [DOI] [PubMed] [Google Scholar]
- 45.Shachar S, Ziv O, Avkin S, Adar S, Wittschieben J, Reissner T, Chaney S, Friedberg EC, Wang Z, Carell T, et al. Two-polymerase mechanisms dictate error-free and error-prone translesion DNA synthesis in mammals. EMBO J. 2009;28:383–393. doi: 10.1038/emboj.2008.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Livneh Z, Ziv O, Shachar S. Multiple two-polymerase mechanisms in mammalian translesion DNA synthesis. Cell Cycle. 2010;9:729–735. doi: 10.4161/cc.9.4.10727. [DOI] [PubMed] [Google Scholar]
- 47.Zhou J, Zhang S, Xie L, Liu P, Xie F, Wu J, Cao J, Ding WQ. Overexpression of DNA polymerase iota (Poliota) in esophageal squamous cell carcinoma. Cancer Sci. 2012;103:1574–1579. doi: 10.1111/j.1349-7006.2012.02309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu F, Tian H, Yue W, Li S, Tian X, Qi L, Zhu Y, Hu W. DNA polymerase iota expression in human lung cancer and its correlation with clinicopathological and prognosis of cancer. Chinese Journal of Cancer Biotherapy. 2009;16:503–506. [Google Scholar]
- 49.Wang M, Devereux TR, Vikis HG, McCulloch SD, Holliday W, Anna C, Wang Y, Bebenek K, Kunkel TA, Guan K, et al. Pol iota is a candidate for the mouse pulmonary adenoma resistance 2 locus, a major modifier of chemically induced lung neoplasia. Cancer Res. 2004;64:1924–1931. doi: 10.1158/0008-5472.can-03-3080. [DOI] [PubMed] [Google Scholar]
- 50.Wang Y, Woodgate R, McManus TP, Mead S, McCormick JJ, Maher VM. Evidence that in xeroderma pigmentosum variant cells, which lack DNA polymerase eta, DNA polymerase iota causes the very high frequency and unique spectrum of UV-induced mutations. Cancer Res. 2007;67:3018–3026. doi: 10.1158/0008-5472.CAN-06-3073. [DOI] [PubMed] [Google Scholar]
- 51.Yang J, Chen Z, Liu Y, Hickey RJ, Malkas LH. Altered DNA polymerase iota expression in breast cancer cells leads to a reduction in DNA replication fidelity and a higher rate of mutagenesis. Cancer Res. 2004;64:5597–5607. doi: 10.1158/0008-5472.CAN-04-0603. [DOI] [PubMed] [Google Scholar]
- 52.Denissenko MF, Pao A, Tang M, Pfeifer GP. Preferential formation of benzo[a]pyrene adducts at lung cancer mutational hotspots in P53. Science. 1996;274:430–432. doi: 10.1126/science.274.5286.430. [DOI] [PubMed] [Google Scholar]
- 53.Chiapperino D, Kroth H, Kramarczuk IH, Sayer JM, Masutani C, Hanaoka F, Jerina DM, Cheh AM. Preferential misincorporation of purine nucleotides by human DNA polymerase η opposite benzo[a]pyrene 7,8-Diol 9,10-epoxide deoxyguanosine adducts. J. Biol. Chem. 2002;277:11765–11771. doi: 10.1074/jbc.M112139200. [DOI] [PubMed] [Google Scholar]
- 54.Zhao B, Wang J, Geacintov NE, Wang Z. Poleta, Polzeta and Rev1 together are required for G to T transversion mutations induced by the (+)- and (−)-trans-anti-BPDE-N2-dG DNA adducts in yeast cells. Nucleic Acids Res. 2006;34:417–425. doi: 10.1093/nar/gkj446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vogel U, Thein N, Moller P, Wallin H. Pharmacological coal tar induces G:C to T:A transversion mutations in the skin of muta mouse. Pharmacol. Toxicol. 2001;89:30–34. doi: 10.1034/j.1600-0773.2001.d01-132.x. [DOI] [PubMed] [Google Scholar]
- 56.Yoon JH, Smith LE, Feng Z, Tang M, Lee CS, Pfeifer GP. Methylated CpG dinucleotides are the preferential targets for G-to-T transversion mutations induced by benzo[a]pyrene diol epoxide in mammalian cells: similarities with the p53 mutation spectrum in smoking-associated lung cancers. Cancer Res. 2001;61:7110–7117. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.