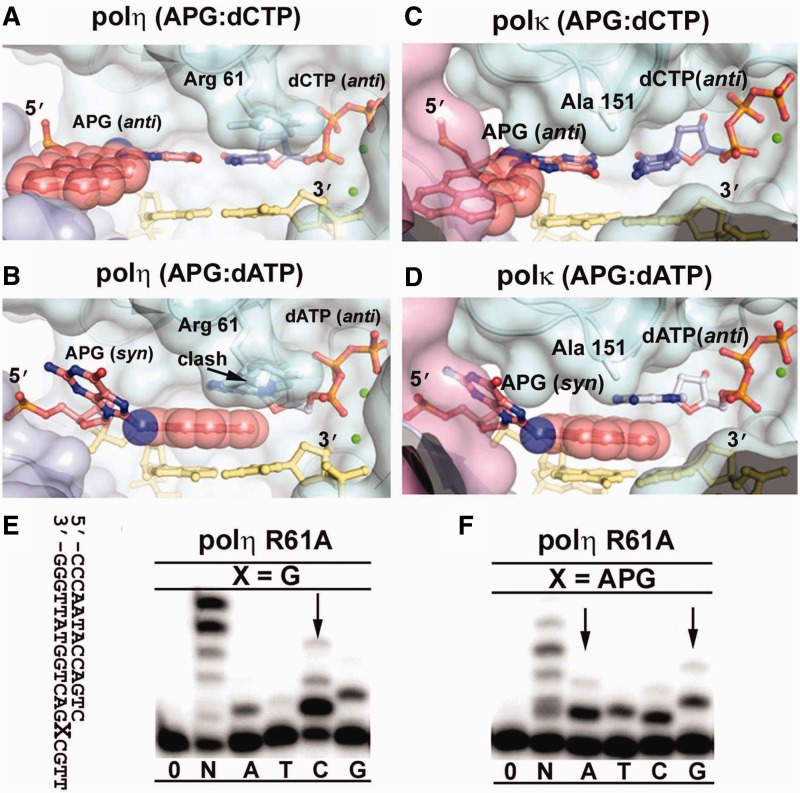
Figure 4.
Modeling of polη and polκ APG replication. The APG:dCTP Watson–Crick base pair (A and C) and the APG:dATP stacking base pair (B and D) from DNA in APG-dCTP and APG-dATP were modeled into polη (3MR2) and polκ(2OH2), respectively. The color scheme is identical to Figure 3. The pink domain in (B and D) is the N-clasp of polκ, which shadows the little finger domain, but does not directly contact DNA. Arg 61 of polη clashing with dATP and Ala 151 of polκ close to dATP are labeled. Replication fidelity of the polη R61A mutant for undamaged G (E) and the APG lesion (F). Polη R61A was incubated with DNA substrates and reacted with no incoming nucleotides (0), all four nucleotides (N) or individual nucleotides (A, T, C, G) for 1 min. The DNA substrate is shown left to the gels. Arrows indicate incoming nucleotide preference to G and increased misinsertion opposite APG.