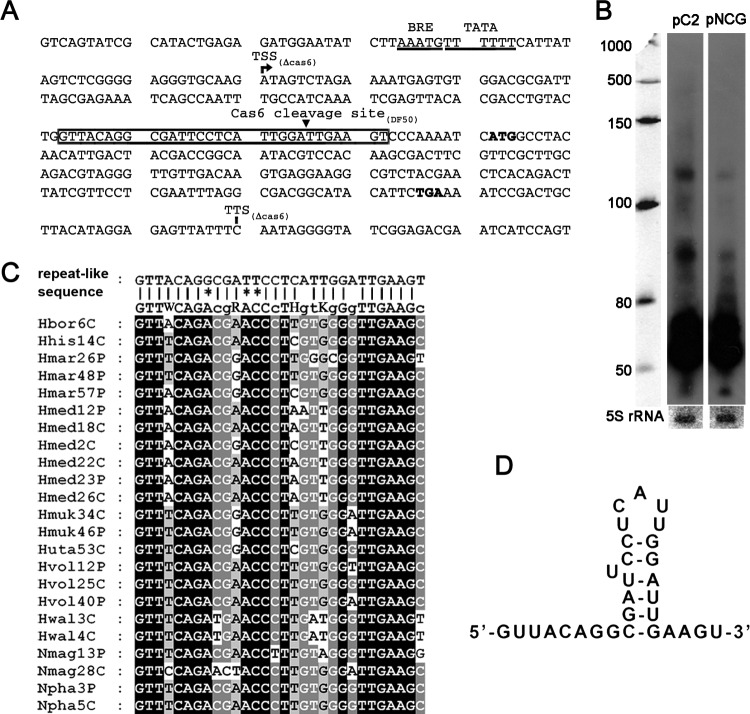
Fig 6.
Analysis of a noncognate repeat-like sequence on the proviral phiH transcript. (A) The repeat-like sequence (boxed) on the phiH transcript. The transcription start (TSS) and termination (TTS) sites determined in a cas6 mutant strain are labeled. The putative BRE and TATA elements are underlined, and the start and stop codons are in bold. The Cas6 cleavage site within the repeat-like sequence determined in the DF50 strain is indicated with a small black triangle. (B) Northern analysis of crRNA production from a modified C2 array (pNCG) whose first repeat has been replaced by the proviral noncognate sequence, with a probe against the spacer sequence. Values to the left indicate RNA size markers, in nucleotides. (C) Alignment of the noncognate repeat of the phiH transcript to the consensus sequence of the haloarchaeal CRISPR repeats (Hbor6C to Npha5C: Hbor, Halogeometricum borinquense; Hhis, Haloarcula hispanica; Hmar, Haloarcula marismortui; Hmed, H. mediterranei; Hmuk, Halomicrobium mukohataei; Hwal, Haloquadratum walsbyi; Hvol, Haloferax volcanii; Huta, Halorhabdus utahensis; Nmag, Natrialba magadii; Npha, Natronomonas pharaonis). Mismatches are indicated by asterisks. P or C represents the location on a plasmid or a chromosome, respectively, and the number denotes the repeat number. Black or grey shading indicates the same nucleotide at this position in all or most sequences, respectively. (D) Secondary structure of the repeat-like RNA sequence predicted by the RNAfold server.