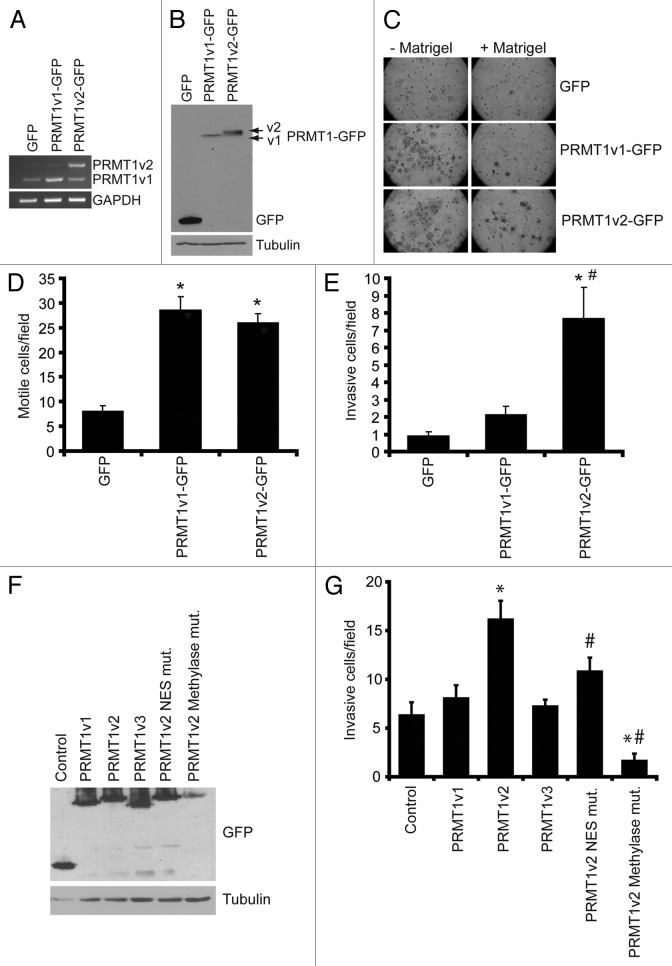
Figure 5. Overexpression of PRMT1v2 in non-invasive breast cancer cells enhances invasion. Total RNA was collected from MCF7 cells stably expressing GFP, GFP-tagged PRMT1v1 (PRMT1v1-GFP) and GFP-tagged PRMT1v2 (PRMT1v2-GFP). PCR analysis of cDNA generated from total RNA using PRMT1 primers (A). GAPDH serves as a loading control. Total protein lysates from MCF7 cells stably expressing GFP, PRMT1v1-GFP or PRMT1v2-GFP were analyzed by western blotting using an anti-GFP antibody (B). Tubulin serves as a loading control. MCF7 stably expressing GFP, PRMT1v1-GFP or PRMT1v2-GFP were analyzed for motility and invasion using Transwell chambers without (- Matrigel) or with a Matrigel layer (+ Matrigel). Cells were plated into Transwell chambers at the same density and the numbers of cells that crossed the chamber membrane was counted after 72 h. Representative images of cells that have passed through the Transwell chamber -/+ Matrigel at 20X magnification (C). Cell numbers that passed through the chamber mebranes without a Matrigel layer (D: motile cells/field) or containing a Matrigel layer (E: invasive cells/field) were counted. Data represents the mean ± standard error of four independent experiments (*p < 0.05 comparing to GFP and #p < 0.05 comparing to PRMT1v1-GFP). MCF7 cells were transiently transfected with plasmid constructs containing a C-terminal EGFP-tagged PRMT1v1, PRMT1v2, PRMT1v3, PRMT1v2 NES mutant or a PRMT1v2 catalytically inactive methylase mutant for 24 h. Western analysis for GFP shows expression of each PRMT1 isoform and PRMT1v2 mutants in MCF7 cells 24 h post transfection (F). To examine invasion, following 24 h transfection, cells were seeded into Matrigel containing Transwell chambers and incubated for 72 h. Cell numbers that passed through the chamber membranes were counted (G). Data represents the mean ± standard error of three independent experiments (*p < 0.05 comparing to control, #p < 0.05 comparing to PRMT1v2).

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.