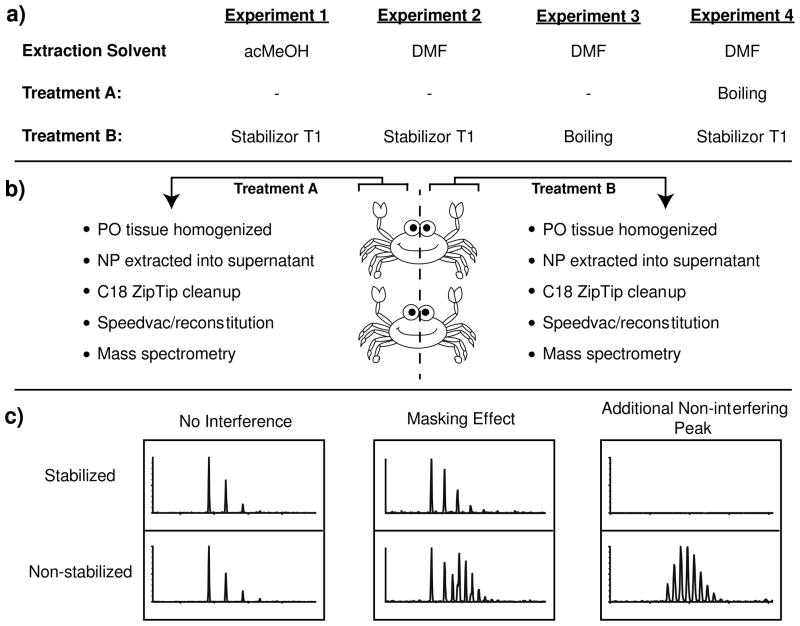
Figure 1.
Experimental workflow. (a) Four experiments were carried out in this study. In experiment 1, acMeOH was used for neuropeptide extraction, whereas in experiments 2, 3, and 4, DMF was used to extract neuropeptides. (a, b) Right and left POs were removed from two crabs. Removed right POs were pooled together and processed using the “Treatment A” workflow. Removed left POs were pooled together and processed using the “Treatment B” workflow. POs were either placed directly into an extraction solvent (acMeOH or DMF) or subjected to tissue stabilization (Stabilizer T1 or boiling) and then placed in extraction solvent. Tissue was then homogenized, neuropeptides extracted, concentrated, and subjected to mass spectral analysis by nanoLC-ESI-MS/MS and MALDI-TOF-MS. (c) It was expected to find instances where no neuropeptide interference would be observed, regions where an isotopic envelope from non-stabilized samples would mask neuropeptide signal, and regions where ion peaks present only in non-stabilized tissues would be detected, constituting additional non-interfering peaks.