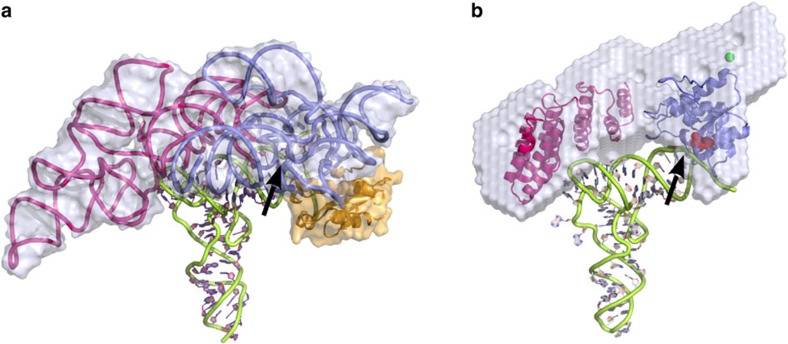
Figure 5. A common mode of RNA binding in RNP and PRORP RNase P.
(a) Structure of Thermotoga maritima ribozyme (PDBid 3Q1R13) with the catalytic domain in blue, the specificity domain in violet, the RNase P protein subunit in orange, the tRNA product in green and the molecular surface of the RNP in grey. (b) Model of A. thaliana PRORP2 protein shown in the same orientation and same colour code: the catalytic domain (NYN domain) in blue, with conserved Asp residues shown as red spheres adjacent to the 5′-cleavage site (indicated by an arrow), and the RNA-binding domain (PPR domain) in violet, interacting with the region of the D-TψC loops, were positioned in the SAXS envelope. The proposed zinc site is indicated by a green sphere in the central region, which connects the two main PRORP domains. This two-domains architecture offers a concave surface, which can be docked on the tRNA acceptor arm. Our data indicate that PRORP proteins have evolved an RNA recognition process very similar to that of RNP RNase P.