Abstract
microRNAs (miRNAs) are essential components of gene regulation, but identification of miRNA targets remains a major challenge. Most target prediction and discovery relies on perfect complementarity of the miRNA seed to the 3′ untranslated region (UTR). However, it is unclear to what extent miRNAs target sites without seed matches. Here, we performed a transcriptome-wide identification of the endogenous targets of a single miRNA—miR-155—in a genetically controlled manner. We found that approximately forty percent of miR-155-dependent Argonaute binding occurs at sites without perfect seed matches. The majority of these non-canonical sites feature extensive complementarity to the miRNA seed with one mismatch. These non-canonical sites confer regulation of gene expression albeit less potently than canonical sites. Thus, non-canonical miRNA binding sites are widespread, often contain seed-like motifs, and can regulate gene expression, generating a continuum of targeting and regulation.
Introduction
miRNAs direct Argonaute (AGO) proteins to post-transcriptionally repress messenger RNA (mRNA) targets and regulate a broad range of physiological processes (Ambros, 2004; Bartel, 2004, 2009). While gain and loss of function studies have established specific roles of individual miRNAs, the identity of most miRNA targets remains unknown, which limits mechanistic insight into observed phenotypes. For instance, it is unclear whether individual miRNAs target multiple components of the same regulatory pathway, individual components of related pathways, or multiple unrelated pathways.
Early studies using both reporter assays and analysis of downregulated genes in miRNA overexpression experiments revealed that perfect complementarity of the 5′ end of the miRNA (i.e. the “seed” region at positions 2-7) to the 3′UTR of target RNAs is the most common determinant of target specificity (Brennecke et al., 2005; Doench and Sharp, 2004; Lewis et al., 2005; Lewis et al., 2003; Lim et al., 2005). However, many reports suggest regulation of sites without perfect seed complementarity (Betel et al., 2010; Brennecke et al., 2005; Didiano and Hobert, 2006; Lal et al., 2009; Lu et al., 2010; Shin et al., 2010; Vella et al., 2004; Vo et al., 2010). Recently, biochemical identification of AGO binding sites became possible with HITS-CLIP and PAR-CLIP techniques, which combine RNase treatment and AGO immunoprecipitation with high-throughput sequencing to identify sites of AGO binding across the transcriptome (Chi et al., 2009; Gottwein et al., 2011; Hafner et al., 2010; Leung et al., 2011; Skalsky et al., 2012; Zisoulis et al., 2010). In these studies, a sizable fraction of CLIP-identified AGO binding sites did not contain seed matches. However, it was unclear whether the majority of this apparently seedless targeting was caused by miRNA independent mechanisms (Leung et al., 2011) or non-canonical miRNA-target interactions. Thus, the mechanism and gene regulatory potential of these seedless interactions remained in question.
To address miRNA dependent targeting by AGO without reliance on sequence motifs, we combined genetic, biochemical, and computational approaches. We analyzed binding sites using differential HITS-CLIP (dCLIP) and mRNA expression changes in primary cells isolated from mice that are wild type or deficient for a single miRNA. We used miR-155 as a model for the following reasons: miR-155 knockout mice show a marked impairment in T and B cell function (Rodriguez et al., 2007; Thai et al., 2007; Vigorito et al., 2007); miR-155 is highly expressed in human malignancies (Eis et al., 2005; Kluiver et al., 2005; Metzler et al., 2004; Volinia et al., 2006) and its overexpression in pre-B cells (Costinean et al., 2006) or hematopoietic stem cells (O’Connell et al., 2008) leads to oncogenic transformation. Importantly, miR-155 is abundant in activated T cells whereas naïve wild type T cells are devoid of miR-155 and are, therefore, similar to miR-155-deficient counterparts. Thus, we employed AGO dCLIP to identify miR-155-dependent binding sites using wild type (WT) and miR-155-deficient (155KO) primary T cells. Our analysis of differential AGO binding confirmed that exact complementarity to nucleotides 2-7 of the miRNA is present in the majority of miR-155-dependent binding sites. We also found that perfect seed matches are absent in ~40% of miR-155-dependent Ago binding sites. These non-canonical sites, which are undetectable by seed-based miRNA target prediction algorithms, were strongly enriched for inexact seed matches, which contained a mismatch to the seed at a single nucleotide position. Furthermore, non-canonical miRNA binding sites regulate gene expression, albeit less potently than canonical sites.
In contrast to seed matches in the 3′UTR, differential gene expression analysis after miRNA perturbation indicated that, as a group, seed matches within coding regions have little gene regulatory activity (Grimson et al., 2007; Lim et al., 2005). Previous studies using CLIP techniques demonstrated extensive AGO binding to the coding region, although these sites were observed to mediate less regulation than 3′UTR sites (Chi et al., 2009; Gottwein et al., 2011; Hafner et al., 2010; Leung et al., 2011; Skalsky et al., 2012; Zisoulis et al., 2010). However, several groups reported features that lead to more regulation by sites in the coding region (Fang and Rajewsky, 2011; Forman et al., 2008; Gu et al., 2009; Schnall-Levin et al., 2011). For this reason, we investigated whether sites with stringently defined Argonaute binding to coding regions lead to functional gene regulation. Using AGO dCLIP we showed miR-155-dependent AGO binding to a subset of miR-155 seed matches within coding regions. However, these sites did not lead to detectable down-regulation of steady state mRNA levels, leaving open the possibility that these sites may have other functions.
Our AGO dCLIP analysis identified, in a genetically controlled manner, a transcriptome-wide set of direct targets for a miRNA in a specific cellular context and revealed the unexpected prevalence of non-canonical targeting by miRNAs.
Results
Analysis of AGO binding sites in miR155-sufficient and -deficient T cells
Although barely detectable in naïve T cells, miR-155 was dramatically upregulated upon activation of CD4+ T cells induced by TCR and co-stimulatory receptor CD28 ligation (Supplementary Figure 1a). In order to reveal miR-155-dependent binding of AGO in primary T cells, we performed AGO dCLIP using activated CD4+ T cells isolated from WT and miR-155 KO C57Bl/6 mice. We UV-cross-linked protein-mRNA complexes in the activated T cells and followed with partial RNase digestion and stringent immunoprecipitation with an anti-Argonaute 2 antibody. The published AGO HITS-CLIP protocol (Chi et al., 2009), was modified to increase coverage and dynamic range and to reduce sequencing costs (see Experimental Procedures).
Libraries constructed from 12 biological replicates of activated 155KO and WT CD4+ T cells were subjected to high-throughput sequencing and the reads were uniquely mapped to the annotated 3′UTR sequences of the mouse genome (a total of 5,213,578 and 5,334,757 uniquely mapping reads from WT and 155KO libraries respectively) (Figure 1a; see Computational Methods). AGO binding sites were detected using a novel peak-calling algorithm based on edge detection (see Supplementary Methods). This algorithm was essential for identifying overlapping binding sites (Supplementary Figure 2b-d) and is effective for binding site detection in other data sets (Supplementary Figure 2e)
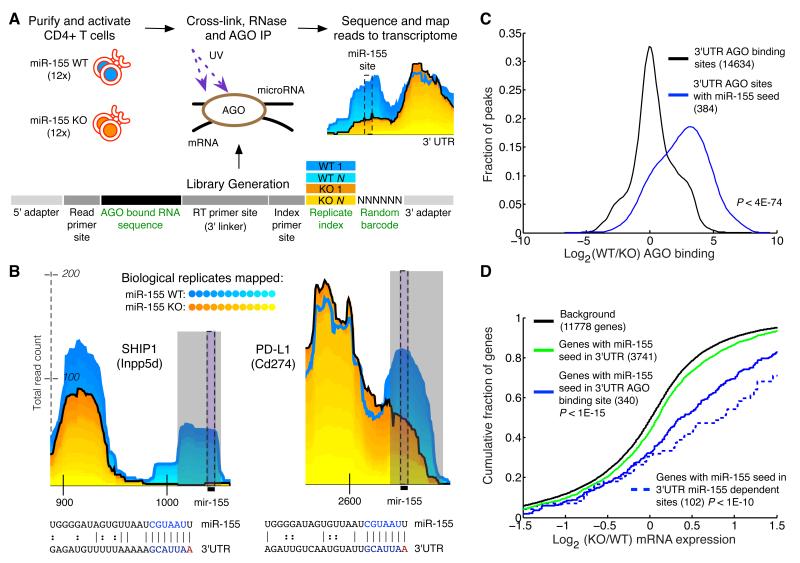
Figure 1. Differential CLIP-sequencing identifies sites of miR-155 mediated Argonaute binding.
(a) CD4+ T cells were isolated from miR-155-sufficient and - deficient mice and RNase treated cell lysates were subjected to Argonaute (AGO) immunoprecipitation. Following stringent purification of complexes, indexed and barcoded libraries were prepared and subjected to Illumina high-throughput sequencing. (b) Examples of AGO binding sites in novel (PD-L1) and established (SHIP1) canonical targets. Grey rectangles indicate miR-155-dependent binding sites (p < .01) called by edge detection method. Reads from the 12 replicates have been stacked; blue shades are miR-155-sufficient (WT) replicates, and yellow shades are mir-155-deficient (155KO) replicates. Black bars indicate the location of the miR-155 seed in the binding site. Coordinates along the x-axis indicate nucleotide position relative to the beginning of the 3′UTR. The y-axis indicates read counts, which are square root transformed following individual library normalization (see Computational Methods). (c) Relative AGO binding in WT and 155KO cells at all 3′UTR binding sites, or 3′UTR binding sites containing a miR-155 seed. (d) Gene expression differences assessed by microarray between WT and 155KO cells of targets predicted by seed, AGO binding sites, or miR-155 dependent AGO binding sites. p-values were calculated by using a one-sided KS test.
In T cells, we identified 14,634 reproducible AGO binding sites in the 3′UTRs of 4,165 genes (Supplementary Table 1 and http://cbio.mskcc.org/leslielab/clipseq). We limited our analysis to these sites, which contained reads mapped from at least seven of the 12 replicates in either WT or 155KO T cell libraries.
Additional analyses confirmed that the observed changes in AGO binding were a direct effect of miR-155 deficiency. First, principal component analysis revealed that the transcriptomes of 155KO and WT activated T cells were very similar (Supplementary Figure 1b). Second, AGO dCLIP libraries from 155KO cells, which included miRNAs, showed no changes in AGO bound miRNA levels other than miR-155 itself (Supplementary Figure 1c). Therefore, miR-155 loss did not cause major changes in the activation state, miRNA expression, or global transcriptome of activated T cells at the time point of analysis.
To validate AGO dCLIP, we examined 3′UTR AGO binding sites containing miR-155 seed matches. We expected these seed match sites to exhibit more AGO binding in WT cells than 155KO cells (Figure 1b). Indeed, for the vast majority of the known targets of miR-155 (Bolisetty et al., 2009; Costinean et al., 2009; Levati et al., 2011; O’Connell et al., 2009; O’Connell et al., 2008; Rai et al., 2010; Wang et al., 2011) we observed differential AGO binding at miR-155 seed matches. Using this approach, we identified many novel targets including PDL1, the ligand for the key inhibitory receptor PD1. Analysis of all AGO sites containing miR-155 seed matches showed that they had significantly reduced binding in 155KO cells compared to all AGO binding sites (p< 4x10-74) (Figure 1c). The observed differential binding at canonical miR-155 targets confirms that AGO dCLIP detects miR-155-dependent binding sites.
We used a stringent statistical test to discern miR-155-dependent sites from other AGO binding sites. At 191 AGO binding sites in 175 genes there was significantly more AGO binding in WT than 155KO cells (cutoff at p < .01; see Computational Methods and Supplementary Table 2). We next investigated whether differential AGO binding correlated with greater target regulation of AGO binding sites containing miR-155 seed matches (57% of all miR-155-dependent sites). Specifically, we compared expression changes in 155KO T cells for genes with AGO binding at miR-155 seed matches against the subset of genes with significant differential AGO binding at miR-155 seed matches (Figure 1d). As expected, we found stronger regulation of target genes with differential AGO binding at miR-155 seed matches. To quantitatively examine whether differential AGO binding is associated with greater gene regulation at AGO bound miR-155 seed matches, we performed a linear regression analysis, which showed that differential AGO binding at sites with miR-155 seed matches correlated with downregulation (p<0.05) (Supplementary Figure 2f). Nearly all (>87%) AGO binding sites containing miR-155 seed matches had less binding in 155KO cells than in WT cells (using a normalized read count difference of 0). Furthermore, genes with AGO bound miR-155 seed matches were greatly induced relative to genes with miR-155 seed matches at which no AGO binding is detected (Figure 1d). Altogether, these data indicate that dCLIP improves the specificity of CLIP, yet for highly expressed miRNAs, presence of a miRNA seed match within an AGO binding site usually identifies a miRNA target.
Analysis of AGO binding sites outside the 3′UTR
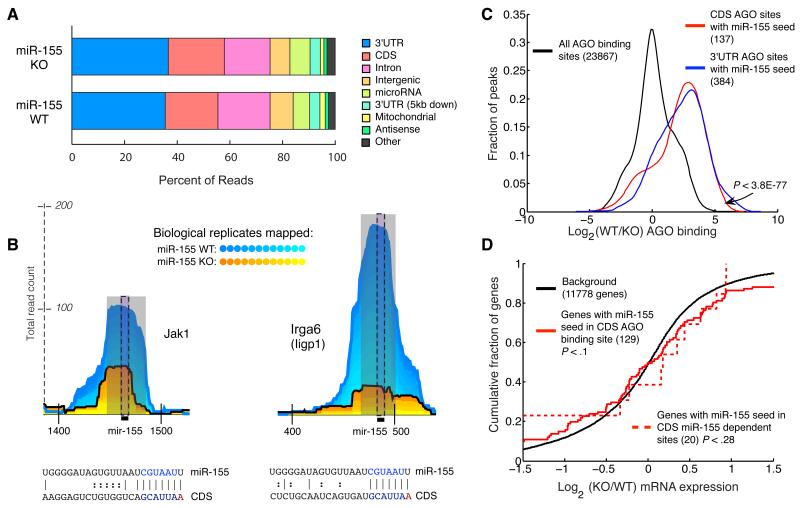
In addition to the prevalent binding that occurs in the 3′UTR, we observed reads mapping to other transcribed regions including the 5′UTR, coding regions, and the introns of genes (Figure 2a). Closer inspection revealed that most intronic reads were unclustered or mapped to snoRNAs. AGO binding to snoRNAs was previously described (Ender et al., 2008; Saraiya and Wang, 2008; Taft et al., 2009), and our experiments provide further data concerning AGO binding to transcripts generated by snoRNA loci in mammals.
Figure 2. Argonaute binding to coding sequence.
(a) Proportion of reads from miR-155-sufficient (WT) and miR-155-deficient (155KO) libraries mapping to various regions of the genome. (b) Examples of AGO binding sites in the coding region. Coordinates along the x-axis indicate nucleotide position relative to the beginning of the transcript. (c) Relative AGO binding in WT and 155KO cells at all binding sites, 3′UTR binding sites containing a miR-155 seed, or CDS binding sites containing a miR-155 seed. (d) Gene expression differences assessed by microarray between WT and 155KO cells of targets predicted by seed and AGO binding sites in the coding region. p-values were calculated using a one-sided KS test.
To determine whether AGO bound coding region sites in a miRNA dependent fashion, we examined sites with miR-155 seed matches. Examining binding sites observed in at least seven of twelve replicates, we observed 137 AGO binding sites containing miR-155 seed matches in the coding regions of 129 genes. Many of these sites had clear miR-155-dependent binding (Figure 2b). Indeed, AGO binding at miR-155 seed matches in the coding region and the 3′UTR was similarly miR-155-dependent (Figure 2c), although there were almost three times as many sites in the 3′UTR. This miR-155 dependence confirms that AGO-miRNA complexes bind to the coding region.
Rare codons 5′ of miRNA binding sites in the coding region contribute to more effective miRNA-mediated regulation (Gu et al., 2009). Consistent with this observation, PAR-CLIP revealed that AGO binding occurs at sites flanked by sequences enriched for rare codons (Hafner et al., 2010). Our data confirms this observation (Supplementary Figure 3). Surprisingly, as was seen in the previous study of AGO binding, rare codons appeared to be more common both 5′ and 3′ of AGO binding sites.
We applied the same stringent statistical test for miR-155 dependence that we applied to 3′UTR sites. This identified 20 genes with sites in the coding region that contained a miR-155 seed and displayed significant miR-155 dependence. However, in contrast to 3′ UTR site-containing mRNAs, these mRNAs were not regulated by miR-155 (Figure 2d). Therefore, although miR-155 AGO complexes bound the coding region, this binding was not sufficient for transcript regulation.
Identification of non-canonical AGO binding sites using AGO dCLIP
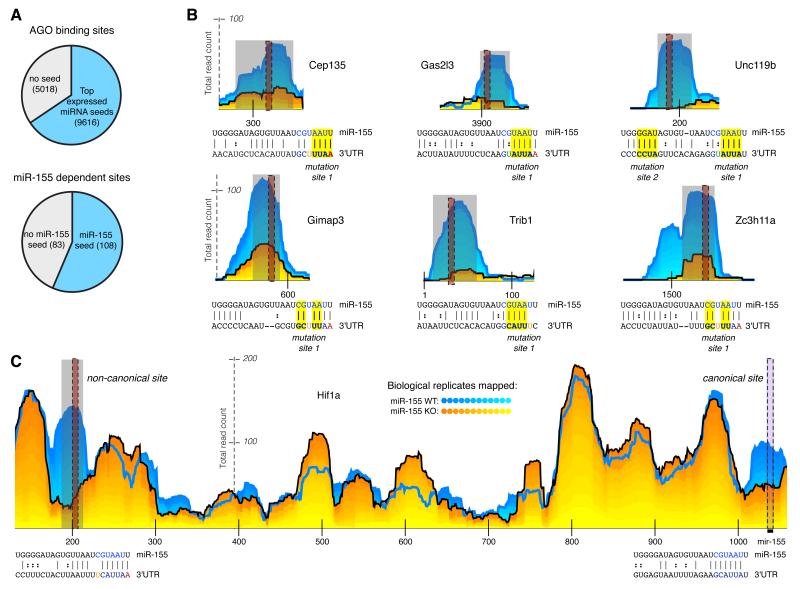
The strength of AGO dCLIP is the ability to identify miRNA dependent binding sites without reliance on seed matches. We found that 34% of 3′UTR AGO binding sites did not have a seed match for any of the 40 highest expressed miRNAs in our cells (Figure 3a and Supplementary Table 3) in agreement with previous reports (Chi et al., 2009; Hafner et al., 2010; Zisoulis et al., 2010). These 40 miRNAs represented more than 75% of the reads mapped to miRNA genes in libraries isolated from WT and 155KO cells (Supplementary Figure 4). Of these miRNAs, the lowest expressed miRNA, had less than 1% of the reads of the highest expressed miRNA. Accounting for AGO binding sites using this many seeds likely overestimates the amount of seed-mediated targeting.
Figure 3. Many miRNA-dependent sites do not contain seed matches.
(a) Proportion of sites from all AGO binding sites or from miR-155-dependent sites that do not contain seed matches. (b-c) Examples of non-canonical targets. Dotted black vertical lines indicate the location of a seed-like site in the site. Predicted base-pairing between miR-155 and 3′UTR sequences are indicated below AGO dCLIP read distribution profiles. Positions selected for mutations in luciferase reporter constructs are highlighted in the alignment. (c) Example of a gene with both a non-canonical and a canonical site in the same 3′UTR; numbers along the horizontal axis indicate nucleotides from the start of the 3′UTR.
Recent work in ES cells suggested that there are miRNA-independent G rich motifs associated with AGO binding sites (Leung et al., 2011); however, these motifs were found in less than 2 percent of AGO binding sites identified in activated T cells. These results suggested that “non-canonical” miRNA dependent targeting is common.
We next focused on miR-155-dependent AGO binding sites in 3′UTRs. While the majority of these sites contained miR-155 seed matches, ~43% did not (Figure 3a). Increasing the stringency for miR-155 dependence (P < .005), did not substantially alter this percentage of seedless sites (~37%). These candidate non-canonical sites without a miR-155 seed, but with significantly more AGO binding in WT vs. 155KO T cells, were found within genes with (e.g., Hif1a and Trib1) and without (e.g., Unc119b, Cep135, Gimap3) miR-155 canonical sites (Figure 3b,c).
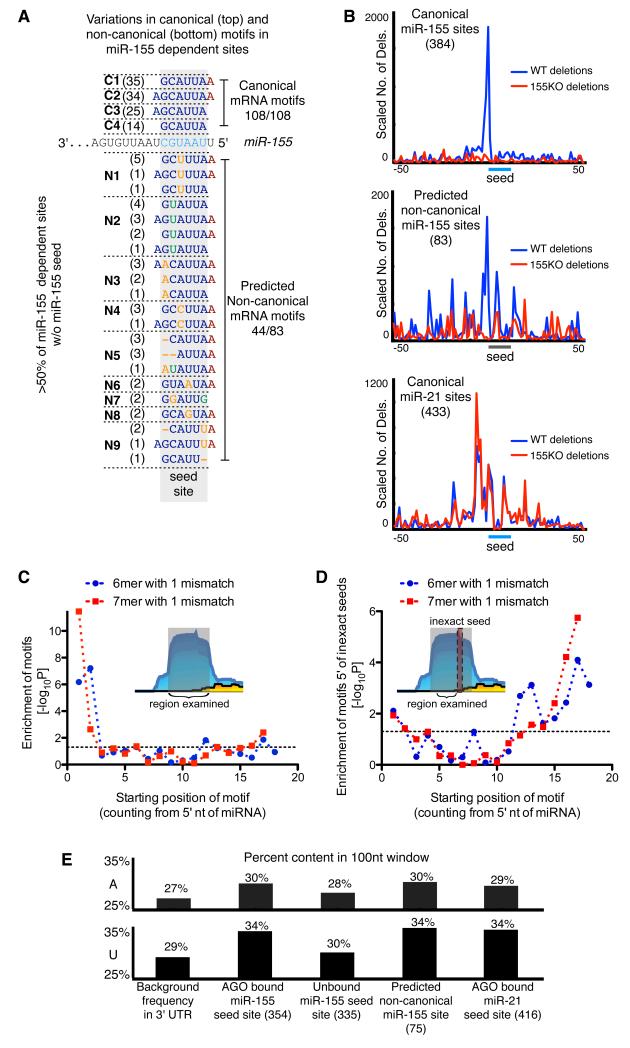
To determine whether miR-155 binds these non-canonical sites directly, we first looked for complementarity to the miR-155 sequence. We implemented a supervised learning strategy to optimize parameters for miR-155/AGO binding site alignments to discriminate between miR-155-dependent binding sites and -independent AGO binding sites (see Supplemental Methods). The optimized parameters rely on extensive complementarity to the seed region of miR-155 to discriminate more than 75% of miR-155-dependent binding sites from non-differential AGO binding sites (average of 10 fold CV; see Supplemental Methods). To identify global patterns in the alignments between miR-155 and miR-155-dependent AGO binding sites, we grouped the alignments based on complementarity to the seed region. The most frequent motifs were represented by sequences with exact complementarity to nucleotides 1-7 (for miR-155 this includes an A at position 1) and nucleotides 1-8. This group of motifs was followed by sequences with complementarity to nucleotides 2-8, and with complementarity to nucleotides 2-7 (Figure 4a). However, in addition to these expected canonical motifs, we found several types of seed-like motifs not previously described at a transcriptome-wide level. These included motifs with inexact complementarity to the miR-155 seed such as mismatches at positions 5 and 7 and a G:U wobble at position 6 (Figure 4a). The observed complementarity for miR-155 within non-canonical AGO binding sites implied that AGO dCLIP specifically identified sites of direct miR-155 targeting.
Figure 4. Characteristics of non-canonical sites.
(a) Variations in canonical (top) and non-canonical (bottom) motifs in miR-155-dependent sites. Number in parentheses indicates the number of miR-155-dependent AGO binding sites containing the motif. (b) Cross-linking induced deletions around canonical seed matches and non-canonical seed-like motifs. (c) Enrichment for inexact 6mer and 7mer motifs in non-canonical miR-155-dependent binding sites relative to all binding sites. (d) Enrichment of 3′ complementarity in non-canonical miR-155 binding sites. Enrichment of motifs complementary to miR-155 was examined in a 16-nucleotide regions found 3-18nt 5′ of inexact seed matches in non-canonical sites relative to equally sized regions in Argonaute binding sites lacking miR-155 seed matches. Enrichments were calculated using Fisher’s Exact Test. (e) AU content in a 100nt window centered at seed matches or non-canonical seed-like motifs.
We next investigated whether these seed-like motifs were able to mediate in vivo interactions with miR-155 loaded AGO complexes. Previous work showed that a small percentage of HITS-CLIP reads contain deletions, which are likely a consequence of reverse transcription errors at sites of UV-induced protein-RNA cross-linking (Zhang and Darnell, 2011). Because UV-mediated cross-linking was preformed on live cells, deletions within binding sites are evidence of in vivo interactions. We mapped deletions surrounding sites of miR-155 canonical motifs and found a large number of deletions proximal to miR-155 seed matches in libraries generated from WT but not 155KO T cells (Figure 4b). As expected, deletions proximal to miR-21 seeds were found equally in WT and 155KO libraries. To assess frequencies of deletions at non-canonical miR-155-dependent AGO binding sites, we used the seed-like motifs identified by our supervised learning algorithm. We found many deletions in WT but not 155KO libraries near seed-like motifs in non-canonical sites. This result indicated that our sequence alignment strategy effectively identified miR-155 binding within AGO bound regions and demonstrated that the interaction between miR-155 and non-canonical sites occurs in vivo.
Features of non-canonical miR-155 binding sites
Further insights into non-canonical sites came from focused analysis of seed-less miR-155-dependent binding sites. These sites exhibited a strong enrichment for seed region motifs that contained a single mismatch from the miRNA seed (up to p<10-10 for the inexact 7mer from nucleotides 1-7) (Figure 4c). A subtle enrichment (p<0.05) for inexact matches to the 3′ end of the miRNA was also apparent. We examined whether non-canonical sites contained sequences complementary to the 3′ end of the miRNA by focusing on sequences 5′ of motifs with a single mismatch from a miR-155 seed, where supplementary 3′ binding is expected to occur. These sequences showed strong enrichment for complementarity to the 3′ end of miR-155 (p<0.03-10-5 for inexact 6mer and 7mer motifs starting at nucleotides 12-18) (Fig 4d), this was a similar enrichment to that seen for canonical sites (Supplementary Figure 5). These results support similar binding for non-canonical and canonical sites with complementarity to the 5′ end of the miRNA and frequent supplementary pairing to the 3′ end of the miRNA.
Since canonical miRNA target sites are flanked with sequences containing a high AU nucleotide content (Grimson et al., 2007; Nielsen et al., 2007), we examined AU content in non-canonical sites (Figure 4e). Consistent with previous studies, we found that sequences flanking AGO-bound miR-155 seed matches displayed higher AU content than unbound miR-155 seed matches and 3′UTRs. Regions around non-canonical sites had higher AU content than 3′UTR sequences or unbound miR-155 sites and in this regard were indistinguishable from AGO bound canonical miR-155 sites.
Non-canonical targeting is associated with moderate regulation of gene expression by miR-155
Our results showed that miR-155 facilitated AGO binding at sites with inexact complementarity to the miR-155 seed. To assess the effect of these seed-like non-canonical sites on gene regulation, we examined targets with a single miR-155 dependent non-canonical site containing a motif with up to one mismatch from positions 1-6 or 2-7 of the miR-155 seed. To conservatively estimate the effect of these sites on gene expression we removed genes with a miR-155 seed match in the 3′UTR. This set of genes was significantly regulated by miR-155 in activated T cells (p<10-4) (Figure 5a). However, these non-canonical targets were less regulated than genes containing a single canonical site and transcripts were not strongly regulated (>2 fold) by a non-canonical target site alone. The regulation of non-canonical targets was confirmed by comparing gene expression of these predicted non-canonical targets to the expression of genes associated with randomly chosen AGO bound sites; this permutation test was also significant (p<0.05). Together this data suggests that non-canonical targeting alone can lead to modest regulation of gene expression.
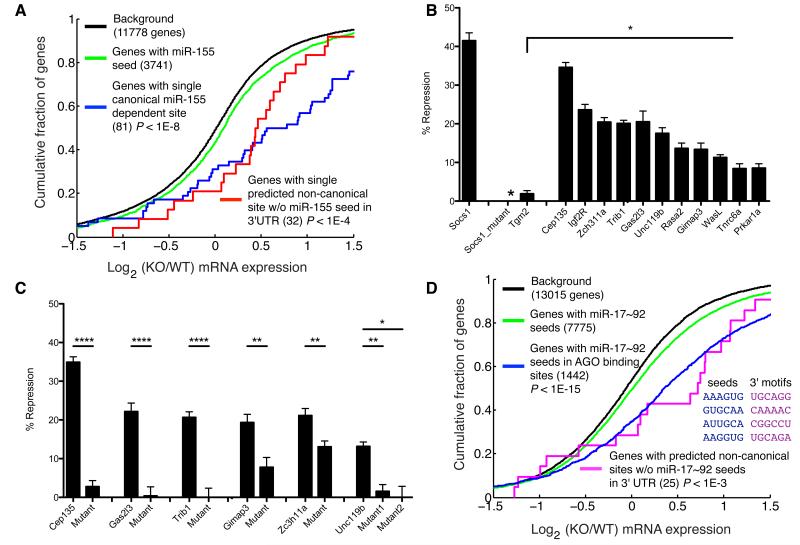
Figure 5. Gene regulation by non-canonical sites.
(a) Gene expression differences assessed by microarray for canonical targets and non-canonical targets without seed matches in the 3′UTR in miR-155-sufficient and -deficient T cells. (b) Repression of 3′UTRs with non-canonical target sites by miR-155 assessed in luciferase reporter assays. Percent repression is the reduction in luciferase activity observed upon miR-155 overexpression relative to the luciferase activity observed with control miRNA overexpression. Socs1, a canonical target, the Socs1 miR-155 site mutant and Tgm2, were used as a positive and negative controls, respectively. (c) Repression of luciferase reporter expression for constructs with and without mutations in the predicted non-canonical sites. Mutations are shown in Figure 3b. (d) Gene expression differences from microarray data (Mu et al., 2009) of canonical targets and non-canonical targets that lacked miR-17~92 seed matches anywhere in the 3′UTR in wild type and miR-17~92 cluster deficient transformed B cells. Predicted non-canonical targets contain AGO binding sites with a single mismatch from miR-17~92 seed matches and an exact match to a 3′ motif. In (b) and (c) mean ± SEM is plotted, *, p<0.05; **, p<0.01; ****, p<10-4.
To test whether miR-155 directly regulates gene expression through binding to non-canonical sites we evaluated the repression of many predicted non-canonical targets using luciferase reporter assays. We generated luciferase reporters containing target sites in their native 3′UTR context (at least 450 nt of the 3′UTR was used) and examined repression by miR-155 overexpressed in HEK293T cells (Figure 5b). Most non-canonical targets were clearly regulated by miR-155. One 3′UTR (Tgm2) with an inexact seed, but without miR-155 dependent AGO binding, was also tested and showed no miR-155 dependent repression. Consistent with the moderate gene expression changes revealed by our global analysis, the non-canonical sites conferred less repression than the canonical target site in the 3′ UTR of the Socs1 gene.
To test whether miR-155 mediated regulation depends on the non-canonical targets sites predicted by AGO dCLIP, we mutated these sites in a subset of luciferase reporter constructs (Figure 5c and Supplementary Figure 9). In most cases we made mutations at four nucleotides in regions with complementarity to the miRNA seed (Figure 3b). In one case, where we predicted contribution of 3′ interactions to miR-155 binding, we also mutated the corresponding 3′ site (Figure 3b). Mutations of the sites significantly reduced repression by miR-155. These experiments demonstrated that miR-155 binding to inexact seed sites can downregulate gene expression.
Prediction of canonical and non-canonical miRNA targets based on gene expression and AGO binding analyses
In addition to a genetically controlled transcriptome-wide identification of miR-155 targets, the AGO dCLIP dataset enables analysis of target interactions with other miRNAs in activated T cells (see Supplementary Table 1). Thus, we investigated whether our datasets were predictive of canonical and non-canonical gene regulation by other endogenous miRNAs. Like miR-155, the miR-17~92 cluster is oncogenic (He et al., 2005), overexpressed in human malignancies (He et al., 2005; Volinia et al., 2006), and regulates differentiation and survival of T and B cells (Jiang et al., 2011; Ventura et al., 2008; Xiao et al., 2008). We used previously published differential gene expression data from B cell lymphoma cells expressing or lacking the miR-17~92 cluster (Mu et al., 2009). Using this dataset, we improved upon seed based predictions by filtering for miR-17~92 seed matches in AGO binding sites (Figure 5d), demonstrating the utility of our dataset for canonical miRNA target identification for other miRNAs and in other cellular contexts. We also observed, moderate induction of genes with inexact complementarity to the miR-17~92 seed (positions 1-6 or 2-7) and exact complementarity to positions 15-20 within AGO binding sites. Importantly, these results were unaffected by exclusion of genes containing miR-17~92 seed matches anywhere in the 3′UTR (Figure 5d). The success of our analysis of miR-17~92 regulation of canonical and non-canonical sites was particularly remarkable considering that we used AGO binding sites identified in activated T cells and the miR-17~92-dependent gene expression changes were measured in transformed B cells.
Analysis of miRNA function in the immune system
Since our dataset provides the most complete and specific list of miRNA targets generated in the immune system, we checked whether targets of individual miRNAs present in activated T cells were enriched for specific immunological functions. Using GO enrichment analysis, we found multiple functions enriched within the targets of miRNAs expressed in our data set (Table 1). Several functions predicted by this analysis have been confirmed; for example, miR-146a is necessary for preventing a T-helper 1 cell-driven autoimmunity (Lu et al., 2010) and miR-17-92 over-expression leads to dysregulated T cell expansion and autoimmunity (Xiao et al., 2008). This analysis would fail unless miRNAs exert their effect through multiple targets. It therefore supports the idea that miRNAs are acting on multiple targets to affect a given function.
Table 1.
GO enrichment analysis. The targets of miRNAs defined by seed matches in AGO CLIP binding sites in T cells were queried for enrichment in Gene Ontology immune processes. All functions with (p<0.005) using a Hypergeometric test are listed.
| miRNA | Immune Process | Targets |
|---|---|---|
| miR-155 | Negative regulation of B cell proliferation | BTLA, INPP5D, RC3H1 |
| miR-155 | Lymphocyte homeostasis | AHR, BCL2, CASP3, HIF1A, MEF2C, PMAIP1, SKIL |
| miR-21 | Alpha-beta T cell differentiation | BCL11B, BCL2, SATB1, STAT6, TCF7 |
| miR-21 | Thymus development | BCL11B, BCL2, BCL2L11, CCNB2, CITED2, JARID2 |
| miR-146 | Enucleate erythrocyte differentiation | ID2, SP1, SP3 |
| miR-146 | Positive regulation of Th1 differentiation | CCR7, SOCS5 |
| miR- 17~92 |
Embryonic hemopoiesis | GATA3, HIF1A, MLL1, RUNX1, TGFBR2 |
| miR- 17~92 |
Negative regulation of T cell proliferation | BTLA, CBLB, CD274, IL2RA, ITCH, PDCD1LG2, RC3H1 |
Based on GO analysis, which implicated miR-155 in lymphocyte homeostasis, we hypothesized that miR-155 might also contribute to expansion of other T cells. We examined expansion of WT and 155KO T cells co-transferred into lymphopenic mice and found that 155KO T cells were outcompeted by WT counterparts (data not shown). This observation was in agreement with an established role for miR-155 in homeostasis of regulatory T cells, a specialized T cell lineage that expresses high levels of miR-155 (Lu et al., 2009). Our analysis demonstrated the power of high-throughput miRNA target discovery for understanding miRNA dependent phenotypes.
Since we identified both the canonical and non-canonical targets of miR-155, we also explored the relationships between these targets (Supplementary Figure 7). We identified several gene networks in which multiple components are targeted by miR-155. Together, these data support the hypothesis that miRNAs exert biological function through targeting of multiple functionally related genes.
Discussion
The analysis of miR-155-dependent AGO binding sites in activated primary T cells provides a transcriptome-wide perspective of a miRNA’s targets in an endogenous context. miR-155-AGO complexes are bound to over 300 canonical sites containing a perfect 6-8mer seed in the 3′UTR and this group of genes is strongly regulated by miR-155.
No miR-155 seeds were observed in ~40% of miR-155 dependent AGO binding sites and the majority of these sites contain inexact seed matches, which are associated with weaker regulation. Several rules for non-canonical sites have been described, notably for bulge sites and centered sites (Chi et al., 2012; Shin et al., 2010). Surprisingly neither bulge sites nor centered sites were found among the miR-155 non-canonical sites. While centered sites are potent regulators of gene expression, they are relatively rare, with an average of several functional sites per miRNA (Shin et al., 2010)—so it was not surprising that they were not observed in our data set. In contrast, bulge sites are relatively common, accounting for up to one quarter of all targets for some miRNAs (Chi et al., 2012); however, miR-155 appears to lack binding to this type of site.
Despite the modest level of regulation associated with non-canonical sites, these sites could play important biological roles. First, modest regulation of many targets may lead to important biological consequences—although there are not yet experimental systems for studying the effects of numerous moderate perturbations in gene expression. In addition, combinations of canonical and non-canonical sites may afford a wide spectrum of regulation of gene expression. Second, recent work suggested that miRNA binding sites on endogenous transcripts can compete for miRNA-AGO complexes and thereby upregulate other genes(Cesana et al., 2011; Jeyapalan et al., 2011; Karreth et al., 2011; Lee et al., 2009; Poliseno et al., 2010; Sumazin et al., 2011; Tay et al., 2011). Third, non-canonical sites may serve as an evolutionary mid-point for stronger canonical miRNA targeting. We therefore examined whether orthologous human 3′UTR sequences of our predicted non-canonical targets were enriched for miR-155 seed matches. We found a modest enrichment (P<.05; hypergeometric test) for canonical motifs in human 3′UTRs, suggesting that non-canonical targets may be an evolutionary mid-point. We also examined conservation using alignments of multiple species (Supplementary Figure 8), which again suggested modest conservation of non-canonical sites.
As mentioned above, recent work suggested a model in which RNAs compete for miRNA-AGO complexes and reduce repression of other RNAs bearing sites for the same miRNAs (Cesana et al., 2011; Jeyapalan et al., 2011; Karreth et al., 2011; Lee et al., 2009; Poliseno et al., 2010; Sumazin et al., 2011; Tay et al., 2011). In this study, we found greater than 500 binding sites for miR-155 in a defined T cell lineage. The most bound site, which occurs in a mitochondrial genome encoded RNA, accounts for ~20% of miR-155 dependent AGO binding. The most bound site transcribed from the nuclear genome accounts for less than 5% of miR-155 dependent AGO binding and only 8 sites can individually account for >1% of miR-155 dependent AGO binding. This suggests that at endogenous transcript levels very few transcripts bind a given miRNA-AGO complex at high enough levels to substantially influence the amount of free complex. Although data supporting competing endogenous RNAs focused on transcripts that share multiple miRNA binding sites, it seems unlikely that a transcript which altered the free pool of multiple miRNAs by <1%, would significantly affect regulation of other targets. Therefore, we propose that if the competing endogenous transcript hypothesis is correct, 1) very few endogenous transcripts bind sufficient miRNA-AGO complexes to alter targeting of other transcripts, or 2) there is an as yet undescribed mechanism by which specific subsets of targets compete for restricted pools of miRNA-AGO complexes.
In summary, AGO dCLIP revealed that non-canonical AGO binding sites are a significant component of miRNA targeting, although they generally exert mild effects on gene regulation. We expect differential CLIP-based techniques will facilitate further identification of functionally relevant non-canonical miRNA targets. It appears that similar to variations in transcription factor binding motifs, there is a spectrum of sequence motifs bound by RISC complexes loaded with a given miRNA and that these variations to the binding sites afford a continuum of miRNA-dependent regulation of gene expression.
Experimental Procedures
Differential HITS-CLIP
CD4+ T cells were harvested from WT and 155KO mice described elsewhere (Thai et al., 2007). T cells were activated by culturing in the presence of CD3 and CD28 antibodies for 4 days at 37°C, 5% CO2. 12 HITS-CLIP libraries were constructed for both the activated WT and 155KO CD4+ cells, 6 with each 3′linker sequence, which enabled statistical modeling of linker-induced biases. The libraries were constructed as described by (Chi et al., 2009) with the following modifications. First, AGO immunoprecipitation was performed with a polyclonal antibody generated against an Argonaute 2 N-terminal peptide (O’Carroll et al., 2007). Second, to reduce contamination between libraries, two different 3′ linker sequences were used and reverse transcription (RT) was performed with primers that distinguish between the 3′ linkers. This permitted selective amplification of libraries. Third, the RT primers contained index tags, which enabled multiplex sequencing of libraries in a single sequencing lane. Fourth, to improve library complexity, the entire cDNA pool from each replicate was amplified in the presence of SYBR green dye using a real-time thermocycler so that amplification could be stopped near the end of the linear phase. Finally, random barcodes in the RT primer were used to distinguish between more frequently cloned regions and fragments preferentially amplified by PCR. We constructed libraries with barcodes six nucleotides in length to extend the previous dynamic range of the assay. Sequencing was done on Illumina GA2x as SE36 reads with an 11nt index read to sequence the index and degenerate barcode.
Gene Expression Analysis
Cells were isolated and cultured as discussed above for 3 days. Cells were resuspended in Trizol, and RNA was isolated according to manufacturer instructions. cDNA libraries were amplified and hybridized to Affymetrix MOE 430A 2.0 chips.
Computational and bioinformatics analyses
We constructed a 3′UTR database for the mouse genome by identifying the longest annotated 3′UTR for each gene in RefSeq or Ensembl. The annotated coding region sequence (CDS) of each gene was selected from the same transcript. We filtered reads to have >20 base quality scores from the 3′ end and mapped uniquely to the mouse genome allowing up to 1 mismatch. Reads that mapped to the same genomic position and contained the same barcode were collapsed to a single read to reduce PCR bias. We also weighted read counts by normalizing to the library size aligned to the 3′UTR database. mRNA expression data was mean centered and unit normalized.
Luciferase Assays
HEK293T cells were cultured at 7× 104 cells/well in a 24-well plate 1 day prior to transfection. psiCheck-2 vector (Promega) containing 3′UTR regions were cotransfected with miR-155-expressing or control miR-146a-expressing pMDH-PGK-EGFP plasmids with Fugene 6 (Roche) in duplicate. Cells were harvested 18 hours later and luciferase activity was assayed using the Dual-Luciferase Reporter Assay System (Promega). Renilla Luciferase (bearing the cloned 3′UTR) activity was normalized to Firefly Luciferase activity. Results from duplicate wells were averaged and multiple (n≥4) independent experiments were pooled. Differences in repression across wild type constructs was assessed using a one-way ANOVA followed by pairwise tests between the Tgm2 negative control and targets, adjusting for multiple comparisons using Dunnett’s criterion. Differences in repression between mutant and wild type constructs were assessed using paired t tests (Prism, GraphPad Software).
Supplementary Material
Highlights.
Differential CLIP-Seq reveals transcriptome-wide sites of miR-155 binding
Many miR-155 binding sites are non-canonical and lack a perfect seed match
Most non-canonical binding sites contain a mismatch to the canonical seed motif
Non-canonical sites mediate gene regulation, albeit weaker than canonical sites
Acknowledgements
We thank E.B. Conn Gantman and A. Mele for help with HITS-CLIP, C. Lee for assistance with sequencing, A. Tarakhovsky for the AGO2 antibody, and L-F. Lu and A. Arvey for helpful discussions. This work was supported by NIH MSTP grant GM07739 (G.B.L.), and NIH grant R37 AI034206 (A.Y.R.). A.Y.R and R.B.D are investigators with the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers # XXXXXXX
Supplementary Website
Accessible binding maps for all 3′UTRs are located at http://cbio.mskcc.org/leslielab/clipseq
References
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolisetty MT, Dy G, Tam W, Beemon KL. Reticuloendotheliosis virus strain T induces miR-155, which targets JARID2 and promotes cell survival. J Virol. 2009;83:12009–12017. doi: 10.1128/JVI.01182-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi SW, Hannon GJ, Darnell RB. An alternative mode of microRNA target recognition. Nat Struct Mol Biol. 2012;19:321–327. doi: 10.1038/nsmb.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costinean S, Sandhu SK, Pedersen IM, Tili E, Trotta R, Perrotti D, Ciarlariello D, Neviani P, Harb J, Kauffman LR, et al. Src homology 2 domain-containing inositol-5-phosphatase and CCAAT enhancer-binding protein beta are targeted by miR-155 in B cells of Emicro-MiR-155 transgenic mice. Blood. 2009;114:1374–1382. doi: 10.1182/blood-2009-05-220814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, Croce CM. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci U S A. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didiano D, Hobert O. Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nat Struct Mol Biol. 2006;13:849–851. doi: 10.1038/nsmb1138. [DOI] [PubMed] [Google Scholar]
- Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, Lund E, Dahlberg JE. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci U S A. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G. A human snoRNA with microRNA-like functions. Mol Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- Fang Z, Rajewsky N. The impact of miRNA target sites in coding sequences and in 3′UTRs. PLoS One. 2011;6:e18067. doi: 10.1371/journal.pone.0018067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman JJ, Legesse-Miller A, Coller HA. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc Natl Acad Sci U S A. 2008;105:14879–14884. doi: 10.1073/pnas.0803230105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwein E, Corcoran DL, Mukherjee N, Skalsky RL, Hafner M, Nusbaum JD, Shamulailatpam P, Love CL, Dave SS, Tuschl T, et al. Viral microRNA targetome of KSHV-infected primary effusion lymphoma cell lines. Cell Host Microbe. 2011;10:515–526. doi: 10.1016/j.chom.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S, Jin L, Zhang F, Sarnow P, Kay MA. Biological basis for restriction of microRNA targets to the 3′ untranslated region in mammalian mRNAs. Nat Struct Mol Biol. 2009;16:144–150. doi: 10.1038/nsmb.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr., Jungkamp AC, Munschauer M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyapalan Z, Deng Z, Shatseva T, Fang L, He C, Yang BB. Expression of CD44 3′-untranslated region regulates endogenous microRNA functions in tumorigenesis and angiogenesis. Nucleic Acids Res. 2011;39:3026–3041. doi: 10.1093/nar/gkq1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S, Li C, Olive V, Lykken E, Feng F, Sevilla J, Wan Y, He L, Li QJ. Molecular dissection of the miR-17-92 cluster’s critical dual roles in promoting Th1 responses and preventing inducible Treg differentiation. Blood. 2011;118:5487–5497. doi: 10.1182/blood-2011-05-355644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karreth FA, Tay Y, Perna D, Ala U, Tan SM, Rust AG, DeNicola G, Webster KA, Weiss D, Perez-Mancera PA, et al. In vivo identification of tumor-suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell. 2011;147:382–395. doi: 10.1016/j.cell.2011.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluiver J, Poppema S, de Jong D, Blokzijl T, Harms G, Jacobs S, Kroesen BJ, van den Berg A. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J Pathol. 2005;207:243–249. doi: 10.1002/path.1825. [DOI] [PubMed] [Google Scholar]
- Lal A, Navarro F, Maher CA, Maliszewski LE, Yan N, O’Day E, Chowdhury D, Dykxhoorn DM, Tsai P, Hofmann O, et al. miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3′UTR microRNA recognition elements. Mol Cell. 2009;35:610–625. doi: 10.1016/j.molcel.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DY, Shatseva T, Jeyapalan Z, Du WW, Deng Z, Yang BB. A 3′-untranslated region (3′UTR) induces organ adhesion by regulating miR-199a* functions. PLoS One. 2009;4:e4527. doi: 10.1371/journal.pone.0004527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AK, Young AG, Bhutkar A, Zheng GX, Bosson AD, Nielsen CB, Sharp PA. Genome-wide identification of Ago2 binding sites from mouse embryonic stem cells with and without mature microRNAs. Nat Struct Mol Biol. 2011;18:237–244. doi: 10.1038/nsmb.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levati L, Pagani E, Romani S, Castiglia D, Piccinni E, Covaciu C, Caporaso P, Bondanza S, Antonetti FR, Bonmassar E, et al. MicroRNA-155 targets the SKI gene in human melanoma cell lines. Pigment Cell Melanoma Res. 2011;24:538–550. doi: 10.1111/j.1755-148X.2011.00857.x. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Lu LF, Boldin MP, Chaudhry A, Lin LL, Taganov KD, Hanada T, Yoshimura A, Baltimore D, Rudensky AY. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell. 2010;142:914–929. doi: 10.1016/j.cell.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu LF, Thai TH, Calado DP, Chaudhry A, Kubo M, Tanaka K, Loeb GB, Lee H, Yoshimura A, Rajewsky K, Rudensky AY. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity. 2009;30:80–91. doi: 10.1016/j.immuni.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzler M, Wilda M, Busch K, Viehmann S, Borkhardt A. High expression of precursor microRNA-155/BIC RNA in children with Burkitt lymphoma. Genes Chromosomes Cancer. 2004;39:167–169. doi: 10.1002/gcc.10316. [DOI] [PubMed] [Google Scholar]
- Mu P, Han YC, Betel D, Yao E, Squatrito M, Ogrodowski P, de Stanchina E, D’Andrea A, Sander C, Ventura A. Genetic dissection of the miR-17~92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23:2806–2811. doi: 10.1101/gad.1872909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen CB, Shomron N, Sandberg R, Hornstein E, Kitzman J, Burge CB. Determinants of targeting by endogenous and exogenous microRNAs and siRNAs. RNA. 2007;13:1894–1910. doi: 10.1261/rna.768207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Carroll D, Mecklenbrauker I, Das PP, Santana A, Koenig U, Enright AJ, Miska EA, Tarakhovsky A. A Slicer-independent role for Argonaute 2 in hematopoiesis and the microRNA pathway. Genes Dev. 2007;21:1999–2004. doi: 10.1101/gad.1565607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell RM, Chaudhuri AA, Rao DS, Baltimore D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc Natl Acad Sci U S A. 2009;106:7113–7118. doi: 10.1073/pnas.0902636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell RM, Rao DS, Chaudhuri AA, Boldin MP, Taganov KD, Nicoll J, Paquette RL, Baltimore D. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J Exp Med. 2008;205:585–594. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai D, Kim SW, McKeller MR, Dahia PL, Aguiar RC. Targeting of SMAD5 links microRNA-155 to the TGF-beta pathway and lymphomagenesis. Proc Natl Acad Sci U S A. 2010;107:3111–3116. doi: 10.1073/pnas.0910667107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraiya AA, Wang CC. snoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS Pathog. 2008;4:e1000224. doi: 10.1371/journal.ppat.1000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnall-Levin M, Rissland OS, Johnston WK, Perrimon N, Bartel DP, Berger B. Unusually effective microRNA targeting within repeat-rich coding regions of mammalian mRNAs. Genome Res. 2011;21:1395–1403. doi: 10.1101/gr.121210.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin C, Nam JW, Farh KK, Chiang HR, Shkumatava A, Bartel DP. Expanding the microRNA targeting code: functional sites with centered pairing. Mol Cell. 2010;38:789–802. doi: 10.1016/j.molcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalsky RL, Corcoran DL, Gottwein E, Frank CL, Kang D, Hafner M, Nusbaum JD, Feederle R, Delecluse HJ, Luftig MA, et al. The viral and cellular microRNA targetome in lymphoblastoid cell lines. PLoS Pathog. 2012;8:e1002484. doi: 10.1371/journal.ppat.1002484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumazin P, Yang X, Chiu HS, Chung WJ, Iyer A, Llobet-Navas D, Rajbhandari P, Bansal M, Guarnieri P, Silva J, Califano A. An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell. 2011;147:370–381. doi: 10.1016/j.cell.2011.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft RJ, Glazov EA, Lassmann T, Hayashizaki Y, Carninci P, Mattick JS. Small RNAs derived from snoRNAs. RNA. 2009;15:1233–1240. doi: 10.1261/rna.1528909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, Karreth F, Poliseno L, Provero P, Di Cunto F, et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147:344–357. doi: 10.1016/j.cell.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- Vella MC, Choi EY, Lin SY, Reinert K, Slack FJ. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′UTR. Genes Dev. 2004;18:132–137. doi: 10.1101/gad.1165404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigorito E, Perks KL, Abreu-Goodger C, Bunting S, Xiang Z, Kohlhaas S, Das PP, Miska EA, Rodriguez A, Bradley A, et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27:847–859. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo NK, Dalton RP, Liu N, Olson EN, Goodman RH. Affinity purification of microRNA-133a with the cardiac transcription factor, Hand2. Proc Natl Acad Sci U S A. 2010;107:19231–19236. doi: 10.1073/pnas.1013162107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HQ, Yu XD, Liu ZH, Cheng X, Samartzis D, Jia LT, Wu SX, Huang J, Chen J, Luo ZJ. Deregulated miR-155 promotes Fas-mediated apoptosis in human intervertebral disc degeneration by targeting FADD and caspase-3. J Pathol. 2011;225:232–242. doi: 10.1002/path.2931. [DOI] [PubMed] [Google Scholar]
- Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, Henderson JM, Kutok JL, Rajewsky K. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Darnell RB. Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data. Nat Biotechnol. 2011;29:607–614. doi: 10.1038/nbt.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zisoulis DG, Lovci MT, Wilbert ML, Hutt KR, Liang TY, Pasquinelli AE, Yeo GW. Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nat Struct Mol Biol. 2010;17:173–179. doi: 10.1038/nsmb.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.