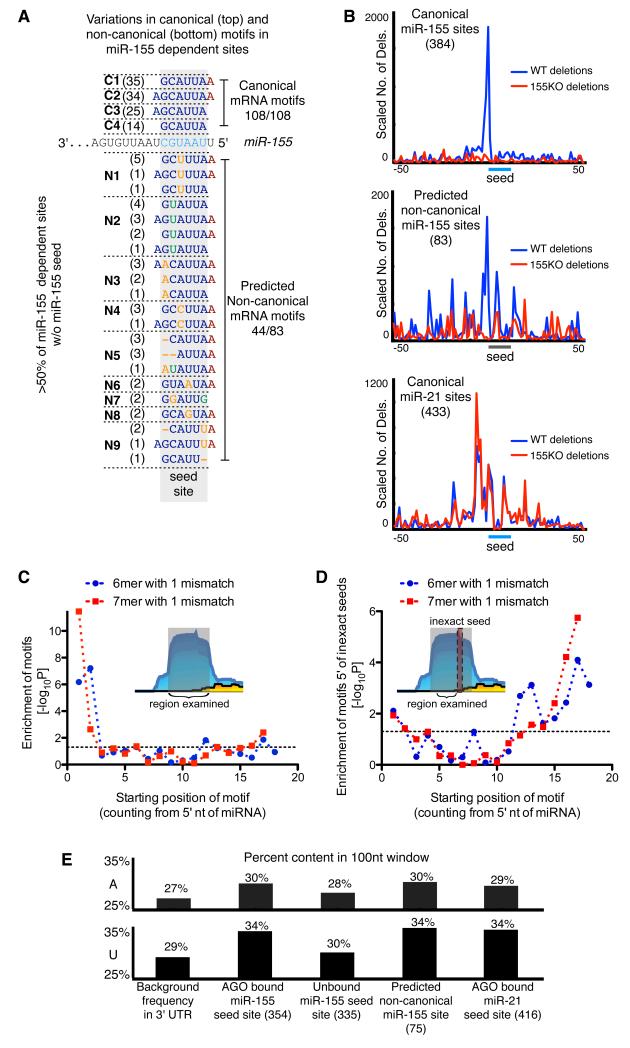
Figure 4. Characteristics of non-canonical sites.
(a) Variations in canonical (top) and non-canonical (bottom) motifs in miR-155-dependent sites. Number in parentheses indicates the number of miR-155-dependent AGO binding sites containing the motif. (b) Cross-linking induced deletions around canonical seed matches and non-canonical seed-like motifs. (c) Enrichment for inexact 6mer and 7mer motifs in non-canonical miR-155-dependent binding sites relative to all binding sites. (d) Enrichment of 3′ complementarity in non-canonical miR-155 binding sites. Enrichment of motifs complementary to miR-155 was examined in a 16-nucleotide regions found 3-18nt 5′ of inexact seed matches in non-canonical sites relative to equally sized regions in Argonaute binding sites lacking miR-155 seed matches. Enrichments were calculated using Fisher’s Exact Test. (e) AU content in a 100nt window centered at seed matches or non-canonical seed-like motifs.