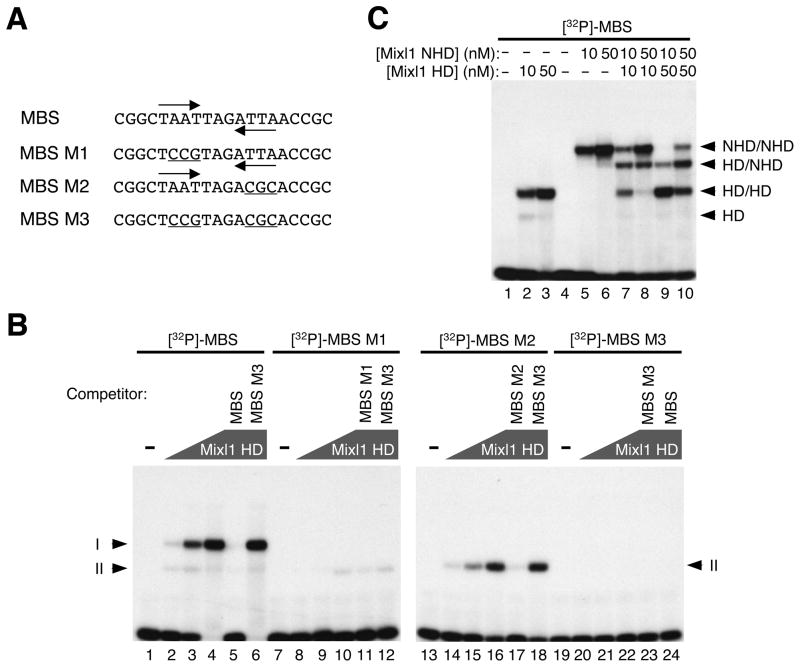
Figure 2. Mixl1 binds the MBS preferentially as a dimer.
(A) Oligonucleotides containing the MBS or one of its mutant forms (MBS M1, MBS M2, and MBS M3) served as probes for EMSA (sense strands shown). The TAAT/ATTA half sites are marked by inverted arrows and mutations within the half sites are underlined. (B) EMSA of Mixl1 HD with [32P]-labeled, double-stranded probes containing the MBS, MBS M1, MBS M2, or MBS M3. Lanes 1, 7, 13 and 19 contained no protein. The concentration of Mixl1 HD was 2 nM (lanes 2, 8, 14, and 20), 10 nM (lanes 3, 9, 15, and 21), and 50 nM (lanes 4–6, 10–12, 16–18, and 22–24), respectively. Mixl1 HD and the MBS probe formed two complexes (Iand II). Mutations in either of the half sites (MBS M1 or MBS M2) abolished the formation of complex I. No complexes were formed when both half sites were mutated (MBS M3). Binding of Mixl1 HD to [32P]-labeled MBS, MBS M1 or MBS M2 was efficiently competed by each of the corresponding unlabeled, oligonucleotides (50X excess) (lanes 5, 11, and 17) but not by MBS M3 (lanes 6, 12 and 18). Polygons represent increasing concentrations of Mixl1 HD. The concentration for each of the unlabeled oligonucleotides (competitors; lanes 5, 6, 11, 12, 17, 18, 23 and 24) was 35 nM. (C) [32P]-labeled, double-stranded MBS was incubated with Mixl1 HD (10 and 50 nM, lanes 2 and 3), Mixl1 NHD (10 and 50 nM, lanes 5 and 6), or both (Mixl1 HD: 10 nM, lanes 7 and 8; 50 nM, lanes 9 and 10. Mixl1 NHD: 10 nM, lanes 7 and 9; 50 nM, lanes 8 and 10). Mixl1 HD formed two complexes with the MBS: HD/HD (complex I) and HD (complex II). Mixl1 NHD formed a dominant complex (NHD/NHD) with the MBS. A complex of intermediate mobility, HD/NHD, was formed by co-incubation of Mixl1 HD and Mixl1 NHD with the MBS probe.