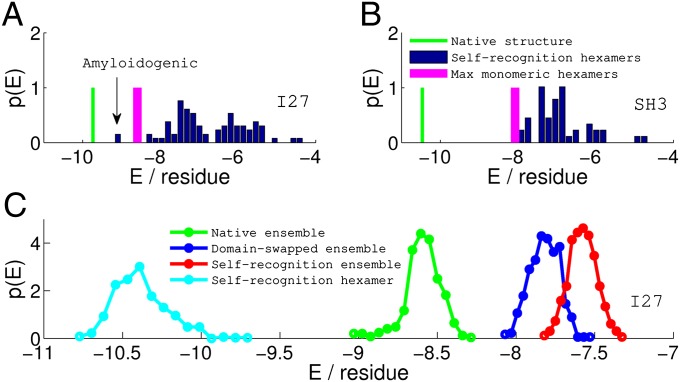
Fig. 5.
Comparisons of stability (energy per residue) among various zero-temperature structures and ensembles of thermally sampled structures of I27 (A and C) and SH3 (B). The stability for the native monomeric structure (green vertical line) is calculated from the AWSEM energy function. The strongest nonnative hexamer pairing possible in the monomer (magenta bar) is significantly less stable than the native structure, indicating that misfolding by inappropriate pairing of strands will be unlikely during the folding of the monomer for both I27 and SH3. In A and B, the blue bars represent the distribution of the stability of all of the self-recognition hexamer pairs, calculated from the AWSEM-Amylometer (SI Text). If the stability of the strongest self hexamer pair is competitive with the native structure, as in the case of I27-I27, the particular self pair becomes responsible for the misfolding of the fused protein in our simulation and potentially, would trigger further aggregation in solution. For SH3-SH3, B predicts that fused protein should fold as well as the monomer in the simulation, because all self hexamer pairings are weaker than the most stable nonnative hexamer pairing in the monomer. In C, the stability distributions of various ensembles of structures collected from the simulations of I27-I27 are shown. The native ensemble (green) is energetically more stable than both the domain-swapped ensemble (blue) and the self-recognition ensemble (red). Nevertheless, the local interactions between the self-pairing hexamers from the self-recognition ensembles, shown in cyan, are even stronger than typical energies in the native folded ensemble.