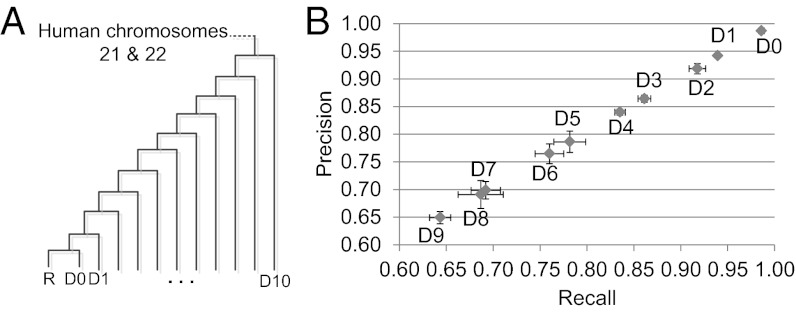
Fig. 2.
Estimation of the accuracy of RACA using simulated genome assemblies. (A) Phylogenetic tree used to generate simulated datasets. Average substitution rates of the generated datasets in comparison with R are D0: 0.059, D1: 0.064, D2: 0.0724, D3: 0.081, D4: 0.090, D5: 0.098, D6: 0.106, D7: 0.114, D8: 0.121, D9: 0.129, D10: 0.136. (B) For all sequence fragments that were generated from the datasets D0–D9, RACA predicted the order and orientation of the sequence fragments by using the dataset R as a reference and more divergent datasets as an outgroup, which were then compared with the true order and orientation. Two evaluation measures were used: (i) recall (x axis), which is the fraction of the true order and orientation of sequence fragments that was found in the predicted sequence fragments, and (ii) precision (y axis), which is the fraction of the predicted order and orientation of sequence fragments that agree with the true order and orientation. For each dataset, the average across five different fragments was displayed, and error bars (horizontal for recall and vertical for precision) represent ±1 SD from the average.