Figure 3.
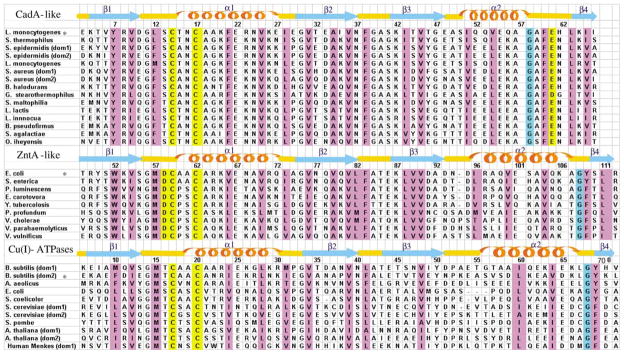
Structure-based sequence alignments of the N termini of various P1-type ATPases. The N-terminal binding domain of CadA from L. monocytogenes, that of ZntA from E. coli and the second domain of CopA from B. subtilis were aligned with other N-terminal binding domains. Secondary structure of NTKII for the CadA-like (this work), ZntA N terminus for the ZntA-like (1MWY) and the second metal-binding site of CopA N terminus for the Cu(I)-ATPases (1JWW) are also shown, together with their amino acid numbering. The asterisk refers to proteins whose secondary structure is shown. Residues that are known or predicted to bind a metal ion are highlighted in yellow. Positions where hydrophobic residues are conserved are highlighted in magenta.
