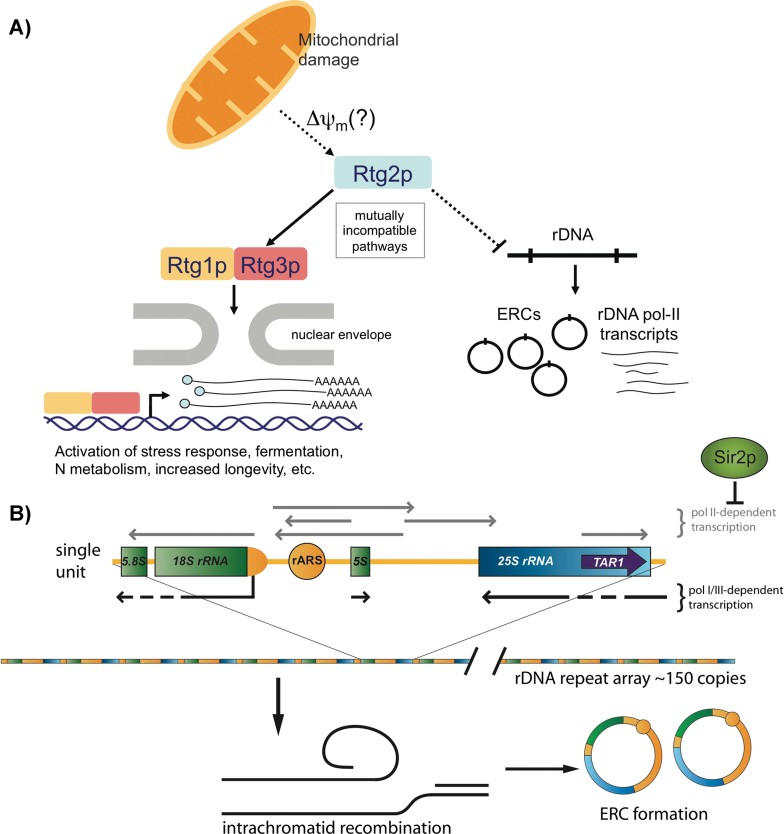
Figure 1.
Relationship between the retrograde response and rDNA. A: Simplified view of the retrograde response. The retrograde response enables yeast cells to detect and respond to mitochondrial damage. In brief, some signal, possibly a drop in mitochondrial membrane potential (Δψm), is transduced by the retrograde response protein, Rtg2p. Rtg2p activates a downstream transcription factor (Rtg1/3p), which leads to activation of suites of genes involved in stress response, nitrogen metabolism, fermentation pathways, and lifespan extension. At the same time, Rtg2p-dependent activation of these processes results in release of Rtg2p-dependent suppression of ERC production/rDNA pol-II silencing. Note that the figure only shows the key features of the retrograde response. Recent reviews on this topic 11, 12 give more detailed descriptions. B: rDNA structure and ERC formation. At the top of the panel, a single rDNA repeat unit is shown. The four ribosomal RNA (rRNA) genes found in yeast are shown as boxes and the spacer regions as orange lines, along with the rARS (rDNA replication origin) and the Tar1 gene antisense to the 25S rRNA gene. The rRNA genes are transcribed by pol-I and -III, and these transcripts are depicted as black arrows below the rDNA unit. A number of pol-II transcripts have also been detected in the rDNA, and these are shown as grey arrows above the rDNA unit. These pol-II rDNA transcripts are normally heavily silenced, with Sir2p thought to play a key role. The tandem-repeat structure of the rDNA is shown beneath the single unit. Formation of ERCs is shown at the bottom of the graphic. During replication, double-strand breaks are repaired by homologous recombination, and if the end pairs with an rDNA unit on the same chromatid (intra-chromatid recombination), a unit (or unit multimer) is ‘popped’ out of the array to form an ERC. ERCs are maintained as they have a replication origin.