Abstract
“Radiomics” refers to the extraction and analysis of large amounts of advanced quantitative imaging features with high throughput from medical images obtained with computed tomography (CT), positron emission tomography (PET) or magnetic resonance imaging (MRI). Importantly, these data are designed to be extracted from standard-of-care images, leading to a very large potential subject pool. Radiomic data are in a mineable form that can be used to build descriptive and predictive models relating image features to phenotypes or gene-protein signatures. The core hypothesis of radiomics is that these models, which can include biological or medical data, can provide valuable diagnostic, prognostic or predictive information. The radiomics enterprise can be divided into distinct processes, each with its own challenges that need to be overcome: (i) image acquisition and reconstruction (ii) image segmentation and rendering (iii) feature extraction and feature qualification (iv) databases and data sharing for eventual (v) ad hoc informatic analyses. Each of these individual processes poses unique challenges. For example, optimum protocols for image acquisition and reconstruction have to be identified and harmonized. Also, segmentations have to be robust and involve minimal operator input. Features have to be generated that robustly reflect the complexity of the individual volumes, but cannot be overly complex or redundant. Furthermore, informatics databases that allow incorporation of image features and image annotations, along with medical and genetic data have to be generated. Finally, the statistical approaches to analyze these data have to be optimized, as radiomics is not a mature field of study. Each of these processes will be discussed in turn, as well as some of their unique challenges and proposed approaches to solve them. The focus of this article will be on images of non-small cell lung cancer, NSCLC.
Keywords: Radiomics, Imaging, Image features, Tumor, Segmentation
1. Introduction
“Radiomics” involves the high throughput extraction of quantitative imaging features with the intent of creating mineable databases from radiological images . It is proposed that such profound analyses and mining of image feature data will reveal quantitative predictive or prognostic associations between images and medical outcomes. In cancer, current radiological practice is generally qualitative; e.g. “a peripherally enhancing spiculated mass in the lower left lobe”. When quantitative, measurements are commonly limited to dimensional measurements of tumor size via one (RECIST) or two (WHO) dimensional long axis measures . These measures do not reflect the complexity of tumor morphology or behavior, nor, in many cases, are changes in these measures predictive of therapeutic benefit . When additional quantitative measures are obtained, they generally average values over an entire region of interest (ROI).
There are efforts to develop a standardized lexicon for the description of such lesions , and to include these descriptors via annotated Image Markup (AIM) into quantitative, mineable data . However, such approaches do not completely cover the range of quantitative features that can be extracted from images, such as texture, shape or margin gradients. In focused studies, texture features have been shown to provide significantly higher prognostic power than ROI-based methods . The modern re-birth of radiomics (or radiogenomics) was articulated in two papers by Kuo and colleagues. Following a complete manual extraction of numerous (>100) image features; a subset of 14 features was able to predict 80% of the gene expression pattern in hepatocellular carcinoma using computed tomographic (CT) images . A similar extraction of features from contrast enhanced (CE) magnetic resonance images (MRI) of glioblastoma was able to predict immunohistochemically identified protein expression patterns . Although paradigm shifting, these analyses were performed manually and the studies were consequently underpowered. In the current iteration of radiomics, image features have to be extracted automatically and with high throughput, putting a high premium on novel machine learning algorithm development.
The goal of radiomics is to convert images into mineable data, with high fidelity and high throughput. The radiomics enterprise can be divided into five processes with definable inputs and outputs, each with its own challenges that need to be overcome: (i) image acquisition and reconstruction; (ii) image segmentation and rendering (iii) feature extraction and feature qualification (iv) databases and data sharing; and (v) ad hoc informatic analyses. Each of these steps must be developed de novo and, as such, poses discrete challenges that have to be met (Figure 1). For example, optimum protocols for image acquisition and reconstruction have to be identified and harmonized. Segmentations have to be robust and involve minimal operator input. Features have to be generated that robustly reflect the complexity of the individual volumes, but cannot be overly complex or redundant. Informatics databases that allow for incorporation of image features and image annotations, along with medical and genetic data have to be generated. Finally, the statistical approaches to analyze these data have to be optimized, as radiomics is not a mature field of study. Variation in results may come from variations in any of these individual processes. Thus, after optimization, another level of challenge is to harmonize and standardize the entire process, while still allowing for improvement and process evolution.
Figure 1.
The process and challenges in radiomics.
2. Image Acquisition and Reconstruction Challenges
In routine clinical image acquisition there is wide variation in imaging parameters such as image resolution (pixel size or matrix size and slice thickness), washout period in the case of PET imaging, patient position, and the variations introduced by different reconstruction algorithms and slice thickness, which are different for each scanner vendor. Even this simple set of imaging issues can create difficulty in comparing results obtained across institutions with different scanners and patient populations. In addition, it is a challenge to identify and curate a large number of image data examples with similar clinical parameters such as disease stage.
2.1 Image Acquisition and Reconstruction
CT
Of all the imaging modalities, CT appears to be the most straightforward and perhaps the easiest to compare across institutions and vendors. Standard phantoms such as the CAT phantom have become the standard of the industry (Figure 2). The phantom is based on the American Association of Physicists in Medicine (AAPM) task group report-1 and has several sections to evaluate imaging performance. There are sections (a) to evaluated the true slice thickness and variation of Hounsfield Units (HUs) with electron density, (b) to look at the ability to visualize small variations in density (low contract delectability), and another for (c) detecting special resolution, high contrast detectability, and a region of uniform medium to examine variation in HUs. The imaging performance of a scanner will depend also on the imaging technique. As the slice thickness is reduced, the photon statistics within a slice are reduced unless the mA or kVp is increased. The axial field of view will also change the voxel size within a slice, and the reconstruction matrix size can also be varied from 512 × 512 up to 1024 × 1024 which also changes the voxel size.
Figure 2.
The computed tomography (CT) phantom. This phantom has several regions to test image quality such as low contrast detectability and spatial resolution.
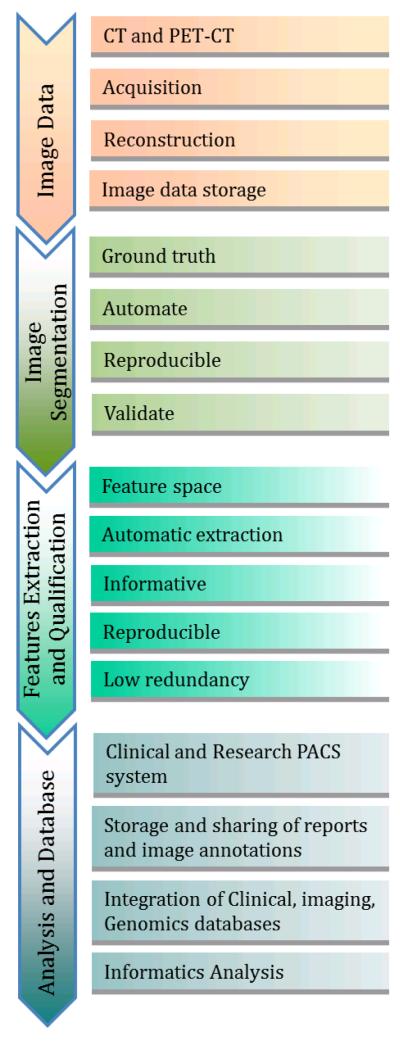
Pitch is a parameter that is frequently optimized by each scanner manufacturer, so that only certain pitches are allowed for an image acquisition. These pitches are unique to each scanner and as a result comparing noise between scanners can only be performed by investigating images acquired using axial, as opposed to helical or spiral, acquisitions. However, helical image acquisitions are used most often in a clinical setting. HUs can also vary with reconstruction algorithm. A single acquisition of a thoracic tumor is shown in Figures 3 A and B, using two different reconstruction algorithms. While this is a single data acquisition, there are significant variations in tumor texture between the two images. The variation in HUs or texture along the vertical paths in Figures 3A and 3B are shown on the graphs (Figures 3C and 3D, respectively).
Figure 3.
Effect of two different reconstruction algorithms on same raw CT data (A and B) where A shows a “standard smooth image” and B shows the same raw data reconstructed using a higher contrast algorithm. To appreciate the effect of these reconstruction algorithms the profiles (in Hounsfield Units) along the vertical lines are shown (C and D, respectively). Even the average Hounsfield Units in the tumor are different for the different algorithms.
For clinical trials, significant effort will be required to match reconstruction protocols and image noise between scanners. While the CAT phantom is a reasonable initial step to compare different scanners, more sophisticated phantoms may be required to match the effects of reconstruction algorithms. Although there can be some variation, different vendors have algorithms that are similar enough to be quantitatively comparable. Indeed the focus of our approach is to use features with a) sufficient dynamic range between patients b) intra-patient reproducibility and c) insensitivity to image acquisition and reconstructions protocol.
PET-CT
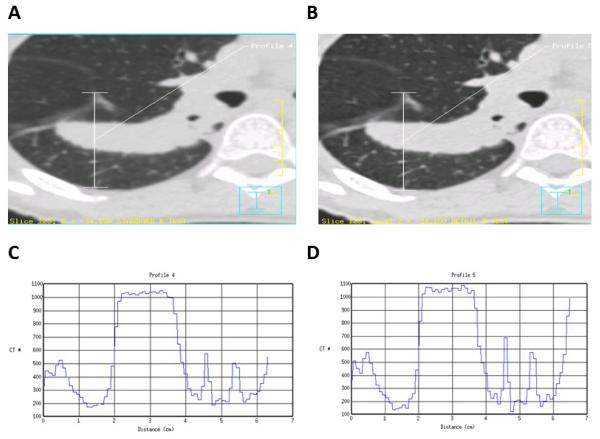
Quantitative imaging with 2-deoxy-2-[18F]fluoro-D-glucose (18-FDG) PET scans is a challenge not only because it requires calibration of the scanner and standardization of the scan protocol but also requires the patient and staff to adhere to a strict patient protocol . From a technical viewpoint the main challenges are the dose calibration and the metabolic volume or VOI reconstruction that depends heavily on the scan protocol and source-to-background ratio . Before a scanner is used in a quantitative manner, inter-institution cross-calibration and quality control such as proposed recently is necessary (Figure 4). From a patient protocol perspective, administration issues (residual activity in syringe, paravenous administration), blood glucose level , uptake period, breathing, patient comfort and inflammation all influence the quantitation of the standardized uptake value (SUV) of 18-FDG. Complying with a strict protocol such as been proposed by the Society of Nuclear Medicine and the European Association of Nuclear Medicine is another prerequisite to quantitative PET imaging.
Figure 4.
Metabolic volume calibration; PET phantom with differently sized sphere sources filled with FDG activity within a background activity. By varying the source to background activity ratio the capability of the PET scanner to reconstruct the correct sphere volume can be quantified.
MRI
The signal intensities in MR images arise from a complex interplay of inherent properties of the tissue, such as relaxation times and acquisition parameters. Therefore, it is difficult to derive information about the physical properties of tissue from MR image signal intensities alone. This is in contrast to CT images where signal intensity can be correlated with the density of the tissue. However, certain techniques, such as diffusion weighted imaging (DWI) and dynamic contrast enhanced (DCE) MRI allow assessment of physiological properties of tissue. For example, the apparent water diffusion coefficient (ADC) determined using DWI varies inversely with tissue cellularity. DCE can be used to extract vascular flow, permeability and volume fractions. Although both of these techniques provide quantitative information, their reliability and reproducibility remain dependent on acquisition parameters and conditions. DWI images can be of low spatial resolution and are sensitive to motion and magnetic susceptibility and the quantitation is dependent on k-space trajectory, gradient strengths, and b-values. DWI has been proposed as a cancer imaging biomarker and there are efforts to develop quality control protocols . Results of the DCE-MRI depend on the contrast agent dose, method of administration, pulse sequence used, field strength of the scanner and the analysis method used . Different investigators use different methods to convert DCE MRI signal intensities to contrast agent concentration . Recently, a group of the Radiological Society of North America (RSNA) known as the Quantitative Imaging Biomarker Alliance (QIBA) initiated a standardization of the protocol for DCEMRI .
Ideally, MR images will all have the same field of view, field strength and slice thickness. Where possible, e.g. brain tumors, multiple sequences with contrast enhancement such as T1-weighted, T2-weighted and FLAIR can be very useful. In magnetic resonance images of human brain tumors, radiomics has the potential to play an important role in categorizing the tumor. It is possible to view the tumor as having different regions using image features, including texture, wavelets, etc. For example, there will be areas of enhancement and potentially necrosis. The tumor bed can be extracted as an expanded region around the post contrast T1 weighted image, for example. Unsupervised clustering can be used to group the data into regions using data from multiple registered sequences. The extraction of image features from those regions, including such things as their location within the tumor bed, can allow for new types of tumor characterization. It has been observed that enhancement in individual tumors can be heterogeneous, and that analysis of this heterogeneity has prognostic value . The location and characteristics of such regions has the potential to provide new insights into tumor prognosis and how well it is likely to respond to targeted treatments. The opportunity to acquire images over time will allow for comparisons and contrasts between regions.
2.2 Need for Large Image Data Sets
The acquisition of images is time-consuming and costly. Because of this, our approach is to focus on standard-of-care images, with the expectation that this will generate large data sets and have more clinical impact, compared to more controlled and dedicated prospective image acquisitions. Radiomics requires large image datasets with the expectation that large numbers may be able to overcome some of the inherent heterogeneities inherent in clinical imaging. Image data sharing across sites will be important to make large datasets available for radiomics analysis.
Various online repositories are available that host image data. The image data contains the image series for each patient and each series containing image slices. One of the largest online CT image repositories is the national biomedical image archive (NBIA) hosted by NCI. Apart from the images, image annotations and outcomes data are also important components to share. There should be a uniform image annotation format which could be read by other users to compare with their own segmentations. This format should support multiple annotations from alternative image analysis algorithms to support higher level processing and prediction. The image data are linked to the metadata in DICOM format images, the metadata contains information about the acquisition, scanner and other details of the images. Currently available clinical image data, which may be used for radiomics study includes LIDC, RIDER and others . Radiomic analyses require refined image data, based on image characteristics (resolution, reconstruction, and acquisition parameters) and clinical parameters (stage of disease, type of disease and outcomes).
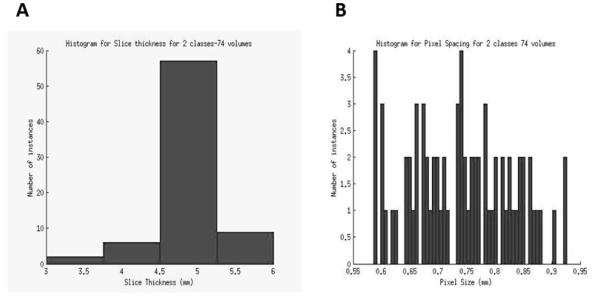
A major use of the information extracted from CT scan images and clinical data is the development of automated prediction models. A challenge in modeling any classifier is making it robust enough for clinical use. Development of robust models requires a sufficiently robust training set. The lack of standardization in imaging makes it difficult to determine the effectiveness of image features being developed and prediction models built to work on those feature values. A snapshot of the extent of the lack of standardization in image acquisition and reconstruction can be seen in the Figure 5. This figure represents the variation in slice thickness and pixel size (in mm) for a dataset of CT-scan images from 74 patients used by Basu et al to develop prediction models for classifying non-small cell lung cancer (NSCLC) tumor types using image features. This variation affects the information being extracted by image feature algorithms, which in turn affects classifier performance. In this scenario, without the presence of a large standardized repository, setting performance benchmarks for effectiveness of image feature algorithms and classifier models built upon those features becomes difficult.
Figure 5.
The variation in slice thickness (A) and pixel size (B) for a dataset of 74 patients.
3. Segmentation Challenges
Segmentation of images into volumes-of-interest (VOI) such as tumor, normal tissue and other anatomical structures is a crucial step for subsequent informatic analyses. Manual segmentation by expert readers is often treated as ground truth; however, it suffers from high inter-reader variability and is labor–intensive; thus not feasible for radiomic analysis requiring very large data sets. Many automatic and semi-automatic segmentation methods have been developed across various image modalities like CT, PET and MRI and also for different anatomical regions like brain, breast, lung, liver etc. Though different image modalities and organ systems require ad hoc segmentation approaches, all share a few common requirements. The segmentation method should be as automatic as possible with minimum operator interaction, time efficient and should provide accurate and reproducible boundaries. Most common segmentation algorithms used for medical images include region-growing based (click- and-grow), level sets and graph cuts. Region growing methods require an operator to select a seed point within the VOI. While these methods are most suitable for relatively homogenous regions, they can be user-dependent and often introduce significant inter-observer variation in the segmentations. We describe here some major challenges came across while developing segmentation methods for NSCLC.
3.1 Challenges in Segmentation of Lung Tumors
The segmentation of CT thorax images usually requires segmentation of lung fields for successive segmentation of lung nodules. Right and left lungs should be automatically segmented which may serve as a preprocessing step. This has been achieved relatively successfully, however, in cases where high intensity tumors are attached to the pleural wall or mediastinum, automatic segmentation often underperforms (Figure 6). In our experience, while using rule-based methods, automatic segmentations often failed in such cases, as evidenced by extension of lung boundaries into the mediastinum or heart.
Figure 6.
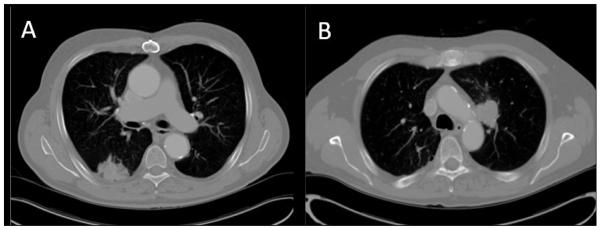
Representative examples of lung tumors attached to anatomical structures like pleural wall, mediastinum or heart that are difficult to segment automatically.
A majority of Stage I and Stage II NSCLC nodules present as homogenous, high-intensity lesions on a background of low-intensity lung parenchyma. These can be segmented with high reproducibility and accuracy. However, partially solid, ground glass opacities (GGOs), nodules attached to vessels and nodules attached to the pleural wall remain difficult to segment automatically and show low reproducibility, especially for Stage III and Stage IV disease. Work is in progress to improve the automatic segmentation and reproducibility in these cases.
A possible solution may come from “crowd-sourcing” the solutions via “segmentation challenges”: public databases for comparing segmentation results via standard metrics. However, there are intellectual property issues that arise from this type of approach. The patentability of an invention stemming from a public database is complicated matter that depends upon a number of factors, including inventorship and the source of funding. In the context of a crowd-sourced development, it may be difficult to identify the “inventors.” It should be noted, however, that there are multiple forms of patent protection– e.g. method patents protecting a particular way of achieving a given result (e.g. an algorithm) or patents covering the particular use of a method. The potential for commercial development may depend only on the resourcefulness of inventors, and the type and scope of the potential patent granted.
Manually traced segmentations are often used as gold standard or ground truth against which the accuracy of the automatic segmentation is evaluated. However, manually traced boundaries themselves suffer from significant inter-reader bias and the reproducibility is low. In a large image dataset, and especially, with slices thickness 3.0 mm or less where number of slices may be higher than 200 per patient, the option of tracing manual boundaries is time prohibitive. Therefore, it is important to have a segmentation algorithm which is automatic and reproducible. The reproducibility of a manual or automatic segmentation of tumors is a known issue. Inter-reader and intra-reader reproducibility significantly varies. As discussed earlier, in radiomics, sources of variations come from acquisition of images, segmentation and analysis, and should be minimized.
3.2 Segmentation Algorithms
Many popular segmentation algorithms have been applied in medical imaging studies within the last 20 years, the most popular ones include region growing methods , level set methods , graph cut methods , active contours (snake) algorithms and semi-automatic segmentations such as livewires , etc.
Region growing algorithms are rapid, but undesired “regions” will be produced if the image contains too much noise. The level set method was initially proposed by Osher and Sethian in 1988 to track moving interfaces and it was subsequently applied across various imaging applications in the late 1990s . By representing a contour as the zero level set of a higher dimensional function (level set function), level set method formulate the motion of the contour as the evolution of the level set function. The graph cut method is relatively new in the area of image segmentation, which constructs an image-based graph and achieves a globally optimal solution of energy minimization functions. Since graph cut algorithms try to identify a global optimum, it is computationally expensive. Another problem for graph cut is the over segmentation.
The active contours (snake) algorithm works like a stretched elastic band being released. The start points are defined around the object which needs to be extracted. The points then move through an iterative process to a point with the lowest energy function value. The active contours requires a good initialization, it is also sensitive to noise, which may lead the snake to undesired locations. The live wire (intelligent scissor) method is motivated by the general paradigm of the active contour algorithm: it converts the segmentation problem into an optimal graph search problem via local active contour analysis and its cost function is minimized by using dynamic programming. A problem with the live wire approach is that it is semi-automatic, requiring multiple human interactions.
There is no universal segmentation algorithm that can work for all medical image applications. With proper parameters settings, each segmentation could segment the region of interest automatically or semi-automatically. However, the result of each segmentation will be quite different and, even for the same algorithm performed multiple times with different initializations, results may be variable. Hence, it is very important to develop agreed-upon metrics to evaluate segmentation algorithms.
3.3. Performance Metrics
Accuracy, reproducibility and consistency are three of the most important factors to evaluate a segmentation algorithm for medical images. However, conventional evaluation metrics normally utilize the manual segmentation provided by radiologists, which is subjective, error-prone and time-consuming. In the majority of cases, manual segmentation tends to overestimate the lesion volume to ensure the entire lesion is identified and the process is highly variable . In other words, “ground truth” segmentation does not exist. Hence, we believe that reproducibility and consistency are more important than accuracy. That is, for a given a tumor, an algorithm must reproducibly provide the same segmentation results that are user-independent.
There is no consensus on the metrics for evaluation of image segmentation algorithms. The metric should address the particular characteristic of the algorithm to be compared, as automated as possible, quantitative and easily computed. Many metrics have been used like volume, center of volume and maximum surface distance, to compare characteristics like robustness and accuracy .
The Jaccard Similarity Index (SI) is the measure of the overlap of two or more volumes and is calculated as the ratio of voxel-wise intersection to union of target and reference images :
| Eq.(1) |
Where, Sa and Sb are segmentations of target and reference images, respectively. An SI of 1.0 represents complete overlap (volume, location and shape) and 0 means no overlap. In our current project, we have calculated SI between each pair of 20 independent computer-generated segmentations of individual lung tumors and report the average SI for each lesion, calculated using following equation:
| Eq.(2) |
Where i ∈ [1, # of cases] is the case index, SIim,in is from Eq. (1). For manual segmentations, the average SI was 0.73. For automated segmentations, the average SI was 0.93.
4. Feature Extraction and Qualification
Once tumor regions are defined, imaging features can be extracted. These features describe characteristics of the tumor intensity histogram (e.g. high or low contrast), tumor shape (e.g. round or spiculated), texture patterns (e.g. homogeneous or heterogeneous), as well as descriptors of tumor location and relations with the surrounding tissues (e.g. near the heart).
4.1. Tumor Intensity Histogram
Tumor intensity histogram-based features reduce the 3-dimensional data of a tumor volume in to a single histogram. This histogram describes the fractional volume for a selected structure for the range of voxel values (e.g. Hounsfield units for a CT scan, or standardized uptake values (SUV) for a FDG-PET scan). From this histogram, common statistics can be calculated (e.g. mean, median, min, max, range, skewness, kurtosis), but also more complex values, such as metabolic volume above an absolute SUV of 5 or the fraction of high density tissue measured with CT . Such threshold values have shown promise in developing classifier models, and optimum thresholds for a given task can be identified with receiver operator characteristic (ROC) analyses. As the outcome (e.g. time to recurrence) to which the threshold is being compared can also have a variable threshold, 3-D ROC approaches have been developed to represent a surface to optimize both the biomarker and the outcome thresholds.
4.2. Shape Based Features
Quantitative features describing the geometric shape of a tumor can also be extracted from the 3-dimensional surface of the rendered volumes . For example the total volume or surface area can be an important characteristic. Also, the surface to volume ratio can be determined, where a speculated tumor has a higher value than a round tumor with a similar volume. Furthermore, descriptors of tumor compactness and shape (spherecity etc.) can also be calculated .
4.3. Texture Based Features
Second order statistics or co-occurrence matrix features can be used for texture classification , and are widely applied in medical pattern recognition tasks . The basis of the co-occurrence features lies on the second-order joint conditional probability density function P(i,j;a,d) of a given texture image. The elements (i,j) of the co-occurrence matrix for the structure of interest represents the number of times that intensity levels i and j occur in two voxels separated by the distance (d) in the direction (a). Here, a matrix can be selected to cover the 26-connected directions of neighboring voxels in 3D space. The matrix size is dependent on the intensity levels within the 3D structure. Subsequently, from this conditional probability density function features can be extracted, e.g. describing autocorrelation, contrast, correlation, cluster prominence, cluster shade, cluster tendency, dissimilarity, energy, homogeneity, maximum probability, sum of squares, sum average, sum variance, sum entropy, or difference entropy, etc. Furthermore, gray level run length features, derived from run length matrices and using run length metrics as proposed by Galloway can be extracted. A gray level run is the length, in number of pixels, of consecutive pixels that have the same gray level value. From the gray level run length matrix features can be extracted describing short and long run emphasis, gray level non-uniformity, run length non uniformity, run percentage, low gray level run emphasis, high gray level run emphasis. As expected, such analyses can generate hundreds of variables, some of which may be redundant. Thus it is important to assess the redundancy of these data using co-variance.
4.4. Feature Qualification
As described above, a dauntingly large number of image features may be computed. However, all these extracted features may not be useful for a particular task. In addition, the numbers of extracted features can be higher than the number of samples in a study, reducing power and increasing the probability of over-fitting the data. Therefore, dimensionality reduction and selection of task-specific features for best performance is a necessary step. Different feature selection methods can be used for this purpose and may exploit machine learning or statistical approaches . Dimensionality reduction can also be achieved by combining or transforming the original features to obtain new set of features by using methods like principal component analysis . In addition to feature selection for informative and non-redundant features, high reproducibility of the features is important in the development of clinical biomarkers, which requires the availability of a test-retest data set.
To reduce the dimensionality of our feature space, we have chosen to combine different ad hoc methods that are agnostically applied to the behavior of the features themselves, prior to evaluating their ability to develop predictive models. Thus, we evaluated features to fulfill three main requirements: highly reproducible, informative and non-redundant. We have applied three methods in serial manner, where the methods were applied successively to select features. The resulting features of one method were used as input to the next. First, using a test-retest lung CT image data set, highly reproducible features were selected based on concordance correlation coefficient, CCC, with a cutoff of 0.85 for high reproducibility. Subsequently, the CCC-prioritized features were analyzed for dynamic range, calculated as the ratio of scalar biological range to the test-retest absolute difference. Features showing high dynamic range considered to be informative. A Dynamic Range of, e.g. 100 can be arbitrarily used as a cut-off, although features with lower dynamic range may also be informative. Finally, the redundancy in the features, selected after passing through reproducibility and dynamic range requirements, can be reduced by identifying highly correlated features based on correlation coefficients across all samples. Correlation coefficients greater than 0.95 are considered to be highly redundant and thus can be combined into a single descriptor. In a test set, the serial application of these three methods was able to reduce a set of 327 quantitative features to between 39 that were reproducible, informative and not redundant. More features could be added by relaxing the Dynamic Range threshold, which was arbitrarily set at 100. These selected features can also be used to develop classifier models based on machine learning algorithms to improve the performance .
5. Databases and Data Sharing
5.1 De-identification
To follow the principle of providing the minimum amount of confidential information (i.e. patient identifiers) necessary to accommodate downstream analysis of imaging data, raw DICOM image data can be stripped of identified headers and assigned a de-identified number. Maintaining de-identified images and clinical data is an important patient privacy safeguard . In the context of DICOM images, Supplement 142 from the DICOM standards committee provides guidance in the process of de-identifying images, including pixel-level data. Software packages, including NBIA, implement these standards. Likewise, molecular data can be de-identified using a similar approach. However, identifiers must be linked between imaging, molecular data and clinical data in order to build classifier models. This can be achieved through IRB approval or through the more expedient use of an “honest broker”. The clinical data are de-identified by removing personal identifiers (including medical record numbers, patient names, social security numbers and addresses) and providing calculated interval-based survival times instead of actual dates which are also personal identifiers. The approach taken within our radiomics effort is to avoid the use of identified imaging or clinical data unless specifically required. This also facilitates the sharing of data within and across institutions since the de-identification occurs at the creation of the dataset.
5.2 RDB: An Integrated Radiomics Database
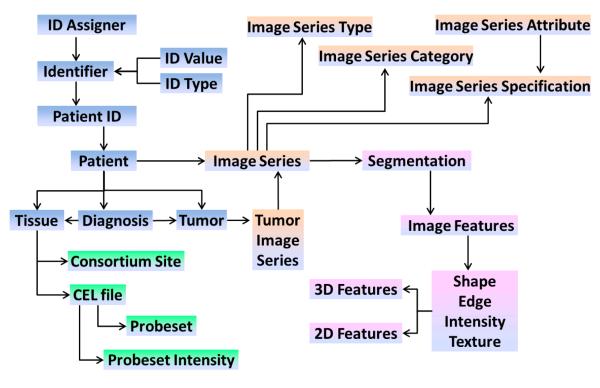
The goal of radiomics is to link the image features to phenotypes or molecular signatures, and this requires development of an integrated database wherein the images and the extracted features are linked to clinical and molecular data (Figure 7). The use of such a database must also be integrated in the workflow starting from image retrieval and calculation of image features up to the joint analysis of image features, clinical and molecular data. Furthermore, as part of a larger network of quantitative imaging sites, we must also be able to exchange data according to an evolving set of standards. Below are some of the challenges discussed in more detail.
Figure 7.
Architecture of the proposed Radiomics database (RDB). High-level database schema capturing the following data types: image types (orange), image features (purple), patient/clinical (blue), and molecular (green) data. Each box represents a set of normalized tables. This schema supports multiple tumors for one patient, with multiple images series, using multiple segmentations generating different image features.
Image Storage
Using clinical PACS (image archive) systems is not amenable for research projects. First, the clinical system is used for operational purposes and introducing additional I/O load and increased storage could negatively impact clinical care. Second, the requirements between research and clinical systems are different and often incompatible. The research image storage server needs to be fully integrated with the downstream data, including molecular and clinical research data. If the imported DICOM images contain Medical Records Numbers (MRN) these need to be linked to other clinical data that are stored on other systems and then the DICOM headers will be de-identified (e.g. patient name). This allows for transparent merging of clinical data across systems. In a research setting, some of the analyses or imaging feature generation software also needs direct access to the DICOM images. Having direct access to the file system where the images are stored makes it possible to create project folders, with all images selected for a specific project, that are specific for the software used for the image feature extraction. In our instance, we are using open source Clear Canvas® as a research PACS system, although others are available.
Integration to Create a Simple Work Stream
In a research setting, it is common that several different software packages are used for image analysis (e.g. 3D-Slicer, Definiens Developer, MITK) and statistical analysis (e.g. R, SAS, Stata). Many of these software packages may be developed by industry, in-house or by other academic groups. This requires that the RDB import data from analysis projects using these software packages in a simple way without sacrificing data integrity. This can be achieved by having the RDB application directly reading working directories and/or results files from the software used. If unique tags have been used when creating image filenames, it is easy to link these data with the right image and downstream clinical and molecular data.
Integration of Clinical and Molecular Data
Integrating data across systems is always a challenge in large settings. The RDB application needs to integrate the data from several systems, such as outcomes and demographic data (Cancer Registry), clinical trial data (e.g. Oncore) or other systems that store clinical and patient information. The manual input of such data should be kept to a minimum through the use of an ETL (extract, transform, and load) tool that captures the physical metadata information to maintain data provenance and minimizes the risk of human errors. The use of a well-developed data dictionary with extensive metadata is essential when integrating data across systems. Therefore, a new data warehouse model that incorporates the metadata layer into the data model, including a comprehensive data dictionary along with calculated data quality attributes such as completeness, accuracy and consistency, has been utilized for the radiomics project . This new data structure was specifically designed to provide easy semantic integration of internal data from multiple heterogeneous source systems as well as provide an easy solution for harmonizing clinical, molecular and imaging data with external members of the quantitative imaging network. Along this path, it has also been important to ensure that the RDB structure and semantics are compatible with those from other institutions and (inter)national databases.
Reporting and Exporting the Data
Advanced statistical analyses of radiomics data require tools such as R, SAS, or MATLAB. The application must be able to export data in such a way that it minimizes any need for processing of data outside the RDB application and thus keeping the data aligned and correct. Longitudinal studies add an extra layer of complexity with the potential need of reporting changes over time, such as imaging features or clinical parameters. A flexible selection of which data should be included and in which format the data should be exported is important.
6. Statistical and Radio-Informatics Analysis
Analysis within radiomics must evolve appropriate approaches for identifying reliable, reproducible findings that could potentially be employed within a clinical context. Applying the existing bioinformatics “toolbox” to radiomic data is an efficient first step since it eliminates the necessity to develop new analytical methods and leverages accepted and validated methodologies. Radiomics-specific analysis issues will exist, as in any field, therefore an important step in achieving consensus on appropriate analysis and evaluation techniques requires availability of real-world data. The goals of the QIN in providing infrastructure to effectively share radiomics data will enable the further development of methodology and best practices within the field.
Some of the more significant methods or developments from the bioinformatics toolbox include: 1) multiple testing issues 2) supervised and unsupervised analysis, and 3) validating biomarker classifiers. Another important analytical consideration is the incorporation of clinical and patient risk factor data since they may have a causal effect or correlation with image features or they may confound statistical associations. Thus, synergizing biostatistics, epidemiology, and bioinformatics approaches is necessary to build robust, parsimonious, and clinically relevant predictive models relating image features to phenotypes/endpoints or gene-protein signatures.
6.1 High-Dimensional Biomarker Discovery and Validation
The field of high-dimensional biomarker discovery and validation has evolved rapidly over the past decade since some of the earliest microarray-based results were reported . In particular, these advances have prompted many studies to address clinical prediction (e.g. prognosis, response to therapy). Many of the lessons learned and tools developed within this field are immediately relevant to the analysis of radiomics datasets.
Multiple Testing
Many of the significant developments within the field of so-called “large-p, small-n” data analysis problems are robust methods for accommodating multiple testing issues. In many datasets in these areas, it is not unusual to test the significance of tens of thousands of variables (p=50,000) using a univariate test (e.g. a t test) across 50 samples (n=50). Any single test may have a low expected false positive rate, however the cumulative effect of many repeated tests guarantees that many statistically significant findings are due to random chance. The false positives (Type I errors in statistics) are controlled using an appropriate p value threshold (e.g p<0.05) in the case of single test. However, performing 50,000 tests creates serious concerns over the accumulated Type I error from such an experiment. This multiple testing problem has been addressed in statistics in many ways however the most familiar, and conservative, Bonferroni corrections severely limit the power of the test in the 50,000-test experiments . False discovery rates (FDR) have been developed to provide more reasonable error estimates. Incorporating this type of correction is an essential step, even in discovery-oriented analysis, to give researchers reasonable guidance on the validity of their discoveries.
Unsupervised and Supervised Data Analysis
Depending on the type of analysis, there are both unsupervised and supervised analysis options available. The distinction in these approaches is that unsupervised analysis does not use any outcome variable, but rather provides summary information and/or graphical representations of the data. Supervised analysis, in contrast, creates models that attempt to separate or predict the data with respect to an outcome or phenotype (for instance, patient outcome or response).
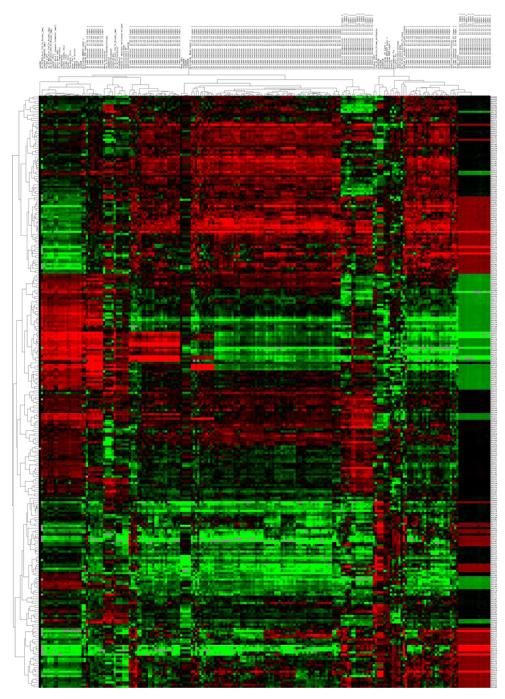
Clustering is the grouping of like data and is one of the most common unsupervised analysis approaches. There are many different types of clustering, although several general types are commonly used within bioinformatics approaches. Hierarchical clustering, or the assignment of examples into clusters at different levels of similarity into a hierarchy of clusters, is the most common type. Similarity is based on correlation (or Euclidean distance) between individual examples or clusters. Most significantly, the data from this type of analysis can be graphically represented using the cluster heat map. Figure 8 represents a heatmap of NSCLC patients with quantitative imaging features extracted. The heat map is an intuitive display that simultaneously reveals row and column hierarchical cluster structure in a data matrix consists of a rectangular tiling with each tile shaded on a color scale to represent the value of the corresponding element of the data matrix. This cluster heat map is a synthesis of various graphic displays developed by statisticians over more than a century .
Figure 8.
Unsupervised hierarchical clustering of lung tumor image features extracted from CT images from 276 non-small cell lung cancer patients. Tumor segmentation for each CT image was performed in a semi-automated fashion. Quantitative imaging features were calculated using Definiens (Munchen, Germany) and represent many 2-dimensional and 3-dimensional characteristics of the tumor. Aspects such as tumor volume, shape and texture were represented. Each of the numerical imaging features was median-centered and all features were clustered using complete linkage, with correlation used as the similarity measure. The resulting heatmap is visualized using red to represent higher than median feature values, and green to represent lower than median feature values. Each row of the heatmap represents a specific imaging feature across patients and each column represents all features for a patient’s lung tumor from CT.
Supervised analysis consists of building a mathematical model of an outcome or response variable. The breadth of techniques available is remarkable and spans statistics and data mining/machine learning. Approaches we have used include neural networks , linear regression and Cox proportional hazards regression . Some essential criteria in selecting an approach include the stability and reproducibility of the model. Neural networks or ensemble methods, if they involve an element of randomness, can lead to results that cannot be replicated without the same random sequences generated. In light of many of the difficulties surrounding genomic-based models, understandability of the generated models is an important consideration. For clinical validation, alternate assays or measurements may be required and thus an understanding of the way in which variables are combined in a decision model is necessary for translation. In the case of NSCLC imaging, methods that generate understandable decisions can be important for combining this information with existing advances in genotyping patients (e.g. EGFR mutation, EML4-ALK rearrangements).
Multivariate data analysis tools such as Principal Component Analysis (PCA) and Partial Least Squares Projection to Latent Structures (PLS) can be used to analyze quantitative features together with additional data. PCA allows for an unsupervised analysis of the data where important features can be extracted and visualized. PCA extracts the underlying structures, Principal Components, so that a high-dimensional space can be visualized in a 2D- or 3D-dimensional space. Additional layers of information can be added by using coloring, shapes and size of the objects on the graphs. PCA can be utilized to find grouping, outliers and other artifacts within the data. To find common underlying structures and correlation between two matrices PLS can be used. PLS has been shown to work well on large and complex data sets with more variables than observations, collinear variables and where there are some missing data.
A final, key contribution from the field of bioinformatics is the approach developed to provide validation of prediction findings from high-dimensional experiments. As was noted in , many genomics-based studies that have been published contain significant analytical errors. These errors compromise the estimates of predictor accuracy or overall findings. Following the best practices in developing and then independently validating the observations in a distinct cohort is essential for reproducible results . For instance, in our radiomics study we have provided for several validation components, including validation between MAASTRO and Moffitt sample sets, as well as validation in prospectively collected Moffitt samples. When model building and cross-validation efforts are completed, the entire group will determine the appropriate model(s) to evaluate in independent validation.
Sample Size Issues
High throughput technologies (CT Images, genomic/proteomic, etc) provide us with enormous amount of multivariate data describing the complex biological process. Ability to predict risks or to draw inferences based on clinical outcomes is bogged by sample size. Efron et al has pioneered the work, studied various cross validation methods and proposed unbiased error estimation called the bootstrap . Renewed interest with genomics technologies has seen estimate errors and issues in small samples in the context of classification . In order to make unbiased inference, synthetic samples were generated following the distribution of the sample groups has been studies in classification and contrasting with popular error estimates .
6.2 Clinical and Risk Factor Data
Incorporating detailed clinical and patient risk factor data into radiomics is important because imaging features may be influenced by patient parameters. Patient parameters may influence the image features via a direct causal association or exert a confounding effect on statistical associations whereby the parameter is correlated with both the independent and dependent variables. For instance, smoking-related lung cancers differ from lung cancers in patients who never smoked and thus smoking status could influence image features, clinical parameters (histology), phenotypes, molecular signatures and endpoints (i.e., survival, recurrence). Addressing the influence of patient parameters in radiomics research by using epidemiologic and biostatistical approaches will minimize spurious relationships by avoiding Type I error. Moreover, predictive models which are more precise and clinically relevant may be developed which target well-characterized and defined patient subgroups rather than a broad heterogeneous disease group. For example, a model that includes all patients with adenocarcinoma of lung would not likely be clinically relevant because of the heterogeneity (biological and clinical) of this histologic subtype. However, a predictive model which focused on adenocarcinoma patients with a specific molecular feature (e.g., EML4-ALK fusion) would likely be informative because of the biological and clinical homogeneity and subsequent targeted therapies. Thus as noted with the bioinformatics “toolbox”, existing epidemiologic and biostatistical approaches can be leveraged towards radiomics research to develop robust and clinically relevant prognostic models, reveal factors that may influence (casually or by confounding) radiomic features, and to explore and mine complex datasets.
Acknowledgement
Radiomics of NSCLC U01 CA143062
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, Stiphout RV, Granton P, Zeghers K, J. GR, Boellard R, Dekker A, Hugo JWLA. Radiomics: extracting more information from medical images using advance feature analysis. The European Journal of Cancer. 2011 doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jaffe CC. Measures of response: RECIST, WHO, and new alternatives. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2006;24(20):3245–3251. doi: 10.1200/JCO.2006.06.5599. [DOI] [PubMed] [Google Scholar]
- 3.Burton A. RECIST: right time to renovate? The Lancet Oncology. 2007;8(6):464–465. [Google Scholar]
- 4.Rubin DL. Creating and curating a terminology for radiology: ontology modeling and analysis. Journal of Digital Imaging. 2008;21(4):355–362. doi: 10.1007/s10278-007-9073-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Opulencia P, Channin DS, Raicu DS, Furst JD. Mapping LIDC, RadLex™, and Lung Nodule Image Features. Journal of Digital Imaging. 2011;24(2):256–270. doi: 10.1007/s10278-010-9285-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Channin DS, Mongkolwat P, Kleper V, Rubin DL. The Annotation and Image Mark-up Project1. Radiology. 2009;253(3):590–592. doi: 10.1148/radiol.2533090135. [DOI] [PubMed] [Google Scholar]
- 7.Rubin DL, Mongkolwat P, Kleper V, Supekar K, Channin DS. Medical imaging on the semantic web: Annotation and image markup. 2008.
- 8.Jackson A, O’Connor JPB, Parker GJM, Jayson GC. Imaging tumor vascular heterogeneity and angiogenesis using dynamic contrast-enhanced magnetic resonance imaging. Clinical Cancer Research. 2007;13(12):3449–3459. doi: 10.1158/1078-0432.CCR-07-0238. [DOI] [PubMed] [Google Scholar]
- 9.Rose CJ, Mills SJ, O’Connor JPB, Buonaccorsi GA, Roberts C, Watson Y, Cheung S, Zhao S, Whitcher B, Jackson A. Quantifying spatial heterogeneity in dynamic contrast-enhanced MRI parameter maps. Magnetic Resonance in Medicine. 2009;62(2):488–499. doi: 10.1002/mrm.22003. [DOI] [PubMed] [Google Scholar]
- 10.Gibbs P, Turnbull LW. Textural analysis of contrast-enhanced MR images of the breast. Magnetic Resonance in Medicine. 2003;50(1):92–98. doi: 10.1002/mrm.10496. [DOI] [PubMed] [Google Scholar]
- 11.Canuto HC, McLachlan C, Kettunen MI, Velic M, Krishnan AS, Neves AA, de Backer M, Hu D, Hobson MP, Brindle KM. Characterization of image heterogeneity using 2D Minkowski functionals increases the sensitivity of detection of a targeted MRI contrast agent. Magnetic Resonance in Medicine. 2009;61(5):1218–1224. doi: 10.1002/mrm.21946. [DOI] [PubMed] [Google Scholar]
- 12.Segal E, Sirlin CB, Ooi C, Adler AS, Gollub J, Chen X, Chan BK, Matcuk GR, Barry CT, Chang HY. Decoding global gene expression programs in liver cancer by noninvasive imaging. Nature biotechnology. 2007;25(6):675–680. doi: 10.1038/nbt1306. [DOI] [PubMed] [Google Scholar]
- 13.Diehn M, Nardini C, Wang DS, McGovern S, Jayaraman M, Liang Y, Aldape K, Cha S, Kuo MD. Identification of noninvasive imaging surrogates for brain tumor gene-expression modules. Proceedings of the National Academy of Sciences. 2008;105(13):5213. doi: 10.1073/pnas.0801279105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boellaard R, O’Doherty MJ, Weber WA, Mottaghy FM, Lonsdale MN, Stroobants SG, Oyen WJ, Kotzerke J, Hoekstra OS, Pruim J, Marsden PK, Tatsch K, Hoekstra CJ, Visser EP, Arends B, Verzijlbergen FJ, Zijlstra JM, Comans EF, Lammertsma AA, Paans AM, Willemsen AT, Beyer T, Bockisch A, Schaefer-Prokop C, Delbeke D, Baum RP, Chiti A, Krause BJ. FDG PET and PET/CT: EANM procedure guidelines for tumour PET imaging: version 1.0. European journal of nuclear medicine and molecular imaging. 2010;37(1):181–200. doi: 10.1007/s00259-009-1297-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boellaard R. Standards for PET image acquisition and quantitative data analysis. Journal of nuclear medicine : official publication, Society of Nuclear Medicine. 2009;50(Suppl 1):11S–20S. doi: 10.2967/jnumed.108.057182. [DOI] [PubMed] [Google Scholar]
- 16.Ollers M, Bosmans G, van Baardwijk A, Dekker A, Lambin P, Teule J, Thimister W, Rhamy A, De Ruysscher D. The integration of PET-CT scans from different hospitals into radiotherapy treatment planning. Radiotherapy and oncology : journal of the European Society for Therapeutic Radiology and Oncology. 2008;87(1):142–146. doi: 10.1016/j.radonc.2007.12.025. [DOI] [PubMed] [Google Scholar]
- 17.Janssen MH, Ollers MC, van Stiphout RG, Riedl RG, van den Bogaard J, Buijsen J, Lambin P, Lammering G. Blood glucose level normalization and accurate timing improves the accuracy of PET-based treatment response predictions in rectal cancer. Radiotherapy and oncology : journal of the European Society for Therapeutic Radiology and Oncology. 2010;95(2):203–208. doi: 10.1016/j.radonc.2010.01.021. [DOI] [PubMed] [Google Scholar]
- 18.Padhani AR, Liu G, Koh DM, Chenevert TL, Thoeny HC, Takahara T, Dzik-Jurasz A, Ross BD, Van Cauteren M, Collins D, Hammoud DA, Rustin GJ, Taouli B, Choyke PL. Diffusion-weighted magnetic resonance imaging as a cancer biomarker: consensus and recommendations. Neoplasia. 2009;11(2):102–125. doi: 10.1593/neo.81328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delakis I, Moore EM, Leach MO, De Wilde JP. Developing a quality control protocol for diffusion imaging on a clinical MRI system. Phys Med Biol. 2004;49(8):1409–1422. doi: 10.1088/0031-9155/49/8/003. [DOI] [PubMed] [Google Scholar]
- 20.Yang X, Knopp MV. Quantifying tumor vascular heterogeneity with dynamic contrast-enhanced magnetic resonance imaging: a review. J Biomed Biotechnol. 2011;2011:732848. doi: 10.1155/2011/732848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galbraith SM, Lodge MA, Taylor NJ, Rustin GJ, Bentzen S, Stirling JJ, Padhani AR. Reproducibility of dynamic contrast-enhanced MRI in human muscle and tumours: comparison of quantitative and semi-quantitative analysis. NMR Biomed. 2002;15(2):132–142. doi: 10.1002/nbm.731. [DOI] [PubMed] [Google Scholar]
- 22.Makkat S, Luypaert R, Sourbron S, Stadnik T, De Mey J. Quantification of perfusion and permeability in breast tumors with a deconvolution-based analysis of second-bolus T1-DCE data. J Magn Reson Imaging. 2007;25(6):1159–1167. doi: 10.1002/jmri.20937. [DOI] [PubMed] [Google Scholar]
- 23.Yang C, Stadler WM, Karczmar GS, Milosevic M, Yeung I, Haider MA. Comparison of quantitative parameters in cervix cancer measured by dynamic contrast-enhanced MRI and CT. Magn Reson Med. 2010;63(6):1601–1609. doi: 10.1002/mrm.22371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Priest AN, Gill AB, Kataoka M, McLean MA, Joubert I, Graves MJ, Griffiths JR, Crawford RA, Earl H, Brenton JD, Lomas DJ, Sala E. Dynamic contrast-enhanced MRI in ovarian cancer: Initial experience at 3 tesla in primary and metastatic disease. Magn Reson Med. 2010;63(4):1044–1049. doi: 10.1002/mrm.22291. [DOI] [PubMed] [Google Scholar]
- 25.McGrath DM, Bradley DP, Tessier JL, Lacey T, Taylor CJ, Parker GJ. Comparison of model-based arterial input functions for dynamic contrast-enhanced MRI in tumor bearing rats. Magn Reson Med. 2009;61(5):1173–1184. doi: 10.1002/mrm.21959. [DOI] [PubMed] [Google Scholar]
- 26.Jackson E, Ashton E, Evelhoch J, Buonocore M, Karczmar G, Rosen M, Purdy D, Gupta S, Zahlmann G. Multivendor, multisite DCE-MRI phantom validation study. 2009.
- 27.Armato SG, 3rd, McLennan G, Hawkins D, Bidaut L, McNitt-Gray MF, Meyer CR, Reeves AP, Zhao B, Aberle DR, Henschke CI, Hoffman EA, Kazerooni EA, MacMahon H, Van Beeke EJ, Yankelevitz D, Biancardi AM, Bland PH, Brown MS, Engelmann RM, Laderach GE, Max D, Pais RC, Qing DP, Roberts RY, Smith AR, Starkey A, Batrah P, Caligiuri P, Farooqi A, Gladish GW, Jude CM, Munden RF, Petkovska I, Quint LE, Schwartz LH, Sundaram B, Dodd LE, Fenimore C, Gur D, Petrick N, Freymann J, Kirby J, Hughes B, Casteele AV, Gupte S, Sallamm M, Heath MD, Kuhn MH, Dharaiya E, Burns R, Fryd DS, Salganicoff M, Anand V, Shreter U, Vastagh S, Croft BY, Clarke LP. The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI): a completed reference database of lung nodules on CT scans. Medical physics. 2011;38(2):915–931. doi: 10.1118/1.3528204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Armato SG, 3rd, Meyer CR, McNitt-Gray MF, McLennan G, Reeves AP, Croft BY, Clarke LP. The Reference Image Database to Evaluate Response to therapy in lung cancer (RIDER) project: a resource for the development of change-analysis software. Clinical pharmacology and therapeutics. 2008;84(4):448–456. doi: 10.1038/clpt.2008.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Basu S, Hall LO, Goldgof DB, Gu Y, Kumar V, Choi J, Gillies RJ, Gatenby RA. Developing a classifier model for lung tumors in CT-scan images. IEEE. 2011:1306–1312. [Google Scholar]
- 30.Stroom J, Blaauwgeers H, van Baardwijk A, Boersma L, Lebesque J, Theuws J, van Suylen RJ, Klomp H, Liesker K, van Pel R, Siedschlag C, Gilhuijs K. Feasibility of pathology-correlated lung imaging for accurate target definition of lung tumors. International journal of radiation oncology, biology, physics. 2007;69(1):267–275. doi: 10.1016/j.ijrobp.2007.04.065. [DOI] [PubMed] [Google Scholar]
- 31.Hojjatoleslami S, Kittler J. Region growing: A new approach. Image Processing, IEEE Transactions on. 1998;7(7):1079–1084. doi: 10.1109/83.701170. [DOI] [PubMed] [Google Scholar]
- 32.Dehmeshki J, Amin H, Valdivieso M, Ye X. Segmentation of pulmonary nodules in thoracic CT scans: a region growing approach. Medical Imaging, IEEE Transactions on. 2008;27(4):467–480. doi: 10.1109/TMI.2007.907555. [DOI] [PubMed] [Google Scholar]
- 33.Sethian JA. Level set methods and fast marching methods: evolving interfaces in computational geometry, fluid mechanics, computer vision, and materials science. Cambridge Univ Pr; 1999. [Google Scholar]
- 34.Malladi R, Sethian JA, Vemuri BC. Shape modeling with front propagation: A level set approach. Pattern Analysis and Machine Intelligence, IEEE Transactions on. 1995;17(2):158–175. [Google Scholar]
- 35.Gao H, Chae O. Individual tooth segmentation from CT images using level set method with shape and intensity prior. Pattern Recognition. 2010;43(7):2406–2417. [Google Scholar]
- 36.Chen YT. A level set method based on the Bayesian risk for medical image segmentation. Pattern Recognition. 2010;43(11):3699–3711. [Google Scholar]
- 37.Krishnan K, Ibanez L, Turner WD, Jomier J. Avila RS. An open-source toolkit for the volumetric measurement of CT lung lesions. Optics Express. 2010;18(14):15256–15266. doi: 10.1364/OE.18.015256. [DOI] [PubMed] [Google Scholar]
- 38.Osher S, Sethian JA. Fronts propagating with curvature-dependent speed: algorithms based on Hamilton-Jacobi formulations. Journal of computational physics. 1988;79(1):12–49. [Google Scholar]
- 39.Boykov Y, Veksler O, Zabih R. Fast approximate energy minimization via graph cuts. Pattern Analysis and Machine Intelligence, IEEE Transactions on. 2001;23(11):1222–1239. [Google Scholar]
- 40.So RWK, Tang TWH, Chung A. Non-rigid image registration of brain magnetic resonance images using graph-cuts. Pattern Recognition. 2011 [Google Scholar]
- 41.Xu N, Bansal R, Ahuja N. Object segmentation using graph cuts based active contours. IEEE. 2003;vol. 42:II-46–53. [Google Scholar]
- 42.Slabaugh G, Unal G. Graph cuts segmentation using an elliptical shape prior. IEEE. 2005:II-1222–1225. [Google Scholar]
- 43.Liu X, Veksler O, Samarabandu J. Graph cut with ordering constraints on labels and its applications. IEEE. 2008:1–8. [Google Scholar]
- 44.Ye X, Beddoe G, Slabaugh G. Automatic graph cut segmentation of lesions in CT using mean shift superpixels. Journal of Biomedical Imaging. 2010;2010:19. doi: 10.1155/2010/983963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu W, Zagzebski JA, Varghese T, Dyer CR, Techavipoo U, Hall TJ. Segmentation of elastographic images using a coarse-to-fine active contour model. Ultrasound in medicine & biology. 2006;32(3):397–408. doi: 10.1016/j.ultrasmedbio.2005.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.He Q, Duan Y, Miles J, Takahashi N. A context-sensitive active contour for 2D corpus callosum segmentation. International journal of biomedical imaging. 2007;2007 doi: 10.1155/2007/24826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen C, Li H, Zhou X, Wong S. Constraint factor graph cut–based active contour method for automated cellular image segmentation in RNAi screening. Journal of microscopy. 2008;230(2):177–191. doi: 10.1111/j.1365-2818.2008.01974.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suzuki K, Kohlbrenner R, Epstein ML, Obajuluwa AM, Xu J, Hori M. Computer-aided measurement of liver volumes in CT by means of geodesic active contour segmentation coupled with level-set algorithms. Medical physics. 2010;37:2159. doi: 10.1118/1.3395579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang L, Li C, Sun Q, Xia D, Kao CY. Active contours driven by local and global intensity fitting energy with application to brain MR image segmentation. Computerized Medical Imaging and Graphics. 2009;33(7):520–531. doi: 10.1016/j.compmedimag.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 50.Mortensen EN, Barrett WA. Interactive segmentation with intelligent scissors. Graphical Models and Image Processing. 1998;60(5):349–384. [Google Scholar]
- 51.Souza A, Udupa JK, Grevera G, Sun Y, Odhner D, Suri N, Schnall MD. Iterative live wire and live snake: new user-steered 3D image segmentation paradigms. 2006:61443N. [Google Scholar]
- 52.Lu K, Higgins WE. Interactive segmentation based on the live wire for 3D CT chest image analysis. International Journal of Computer Assisted Radiology and Surgery. 2007;2(3):151–167. [Google Scholar]
- 53.Lu K, Higgins WE. Segmentation of the central-chest lymph nodes in 3D MDCT images. Computers in biology and medicine. 2011 doi: 10.1016/j.compbiomed.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rexilius J, Hahn HK, Schluter M, Bourquain H, Peitgen HO. Evaluation of accuracy in MS lesion volumetry using realistic lesion phantoms. Acad Radiol. 2005;12(1):17–24. doi: 10.1016/j.acra.2004.10.059. [DOI] [PubMed] [Google Scholar]
- 55.Tai P, Van Dyk J, Yu E, Battista J, Stitt L, Coad T. Variability of target volume delineation in cervical esophageal cancer. Int J Radiat Oncol Biol Phys. 1998;42(2):277–288. doi: 10.1016/s0360-3016(98)00216-8. [DOI] [PubMed] [Google Scholar]
- 56.Cooper JS, Mukherji SK, Toledano AY, Beldon C, Schmalfuss IM, Amdur R, Sailer S, Loevner LA, Kousouboris P, Ang KK, Cormack J, Sicks J. An evaluation of the variability of tumor-shape definition derived by experienced observers from CT images of supraglottic carcinomas (ACRIN protocol 6658) Int J Radiat Oncol Biol Phys. 2007;67(4):972–975. doi: 10.1016/j.ijrobp.2006.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jameson MG, Holloway LC, Vial PJ, Vinod SK, Metcalfe PE. A review of methods of analysis in contouring studies for radiation oncology. Journal of medical imaging and radiation oncology. 2010;54(5):401–410. doi: 10.1111/j.1754-9485.2010.02192.x. [DOI] [PubMed] [Google Scholar]
- 58.Warfield SK, Zou KH, Wells WM. Simultaneous truth and performance level estimation (STAPLE): an algorithm for the validation of image segmentation. Medical Imaging, IEEE Transactions on. 2004;23(7):903–921. doi: 10.1109/TMI.2004.828354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zijdenbos AP, Dawant BM, Margolin RA, Palmer AC. Morphometric analysis of white matter lesions in MR images: method and validation. Medical Imaging, IEEE Transactions on. 1994;13(4):716–724. doi: 10.1109/42.363096. [DOI] [PubMed] [Google Scholar]
- 60.Holub O, Ferreira ST. Quantitative histogram analysis of images. Computer physics communications. 2006;175(9):620–623. [Google Scholar]
- 61.El Naqa I, Grigsby P, Apte A, Kidd E, Donnelly E, Khullar D, Chaudhari S, Yang D, Schmitt M, Laforest R. Exploring feature-based approaches in PET images for predicting cancer treatment outcomes. Pattern recognition. 2009;42(6):1162–1171. doi: 10.1016/j.patcog.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.O’Sullivan F, Roy S, O’Sullivan J, Vernon C, Eary J. Incorporation of tumor shape into an assessment of spatial heterogeneity for human sarcomas imaged with FDG-PET. Biostatistics. 2005;6(2):293–301. doi: 10.1093/biostatistics/kxi010. [DOI] [PubMed] [Google Scholar]
- 63.Jain AK. Fundamentals of digital image processing. Prentice-Hall, Inc.; 1989. [Google Scholar]
- 64.Lam SWC. Texture feature extraction using gray level gradient based co-occurence matrices. IEEE. 1996;vol. 261:267–271. [Google Scholar]
- 65.Haralick RM, Shanmugam K, Dinstein IH. Textural features for image classification. Systems, Man and Cybernetics, IEEE Transactions on. 1973;3(6):610–621. [Google Scholar]
- 66.Galloway MM. Texture analysis using gray level run lengths. Computer graphics and image processing. 1975;4(2):172–179. [Google Scholar]
- 67.Castellano G, Bonilha L, Li LM, Cendes F. Texture analysis of medical images. Clinical radiology. 2004;59(12):1061–1069. doi: 10.1016/j.crad.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 68.Zinovev D, Raicu D, Furst J, Armato SG., III Predicting Radiological Panel Opinions Using a Panel of Machine Learning Classifiers. Algorithms. 2009;2(4):1473–1502. [Google Scholar]
- 69.Soh LK, Tsatsoulis C. Texture analysis of SAR sea ice imagery using gray level co-occurrence matrices. Geoscience and Remote Sensing, IEEE Transactions on. 1999;37(2):780–795. [Google Scholar]
- 70.Suárez J, Gancedo E, Álvarez JM, Morán A. Optimum compactness structures derived from the regular octahedron. Engineering Structures. 2008;30(11):3396–3398. [Google Scholar]
- 71.Tang X. Texture information in run-length matrices. Image Processing, IEEE Transactions on. 1998;7(11):1602–1609. doi: 10.1109/83.725367. [DOI] [PubMed] [Google Scholar]
- 72.Kramer K, Goldgof DB, Hall LO, Remsen A. Increased classification accuracy and speedup through pair-wise feature selection for support vector machines. IEEE. 2011:318–324. [Google Scholar]
- 73.Song F, Guo Z, Mei D. Feature Selection Using Principal Component Analysis. Yichang. IEEE. 2010:27–30. [Google Scholar]
- 74.Heshmati A, Amjadifard R, Shanbehzadeh J. ReliefF-Based Feature Selection for Automatic Tumor Classification of Mammogram Images. Tehran. IEEE. 2011:1–5. [Google Scholar]
- 75.Jain A, Zongker D. Feature selection: Evaluation, application, and small sample performance. Pattern Analysis and Machine Intelligence, IEEE Transactions on. 1997;19(2):153–158. [Google Scholar]
- 76.Fu J, Lee SK, Wong STC, Yeh JY, Wang AH, Wu H. Image segmentation feature selection and pattern classification for mammographic microcalcifications. Computerized Medical Imaging and Graphics. 2005;29(6):419–429. doi: 10.1016/j.compmedimag.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 77.Liu J, Erdal S, Silvey SA, Ding J, Riedel JD, Marsh CB, Kamal J. Toward a fully de-identified biomedical information warehouse. AMIA Annual Symposium proceedings / AMIA Symposium AMIA Symposium; 2009. pp. 370–374. [PMC free article] [PubMed] [Google Scholar]
- 78.Freymann JB, Kirby JS, Perry JH, Clunie DA, Jaffe CC. Image Data Sharing for Biomedical Research-Meeting HIPAA Requirements for De-identification. Journal of digital imaging : the official journal of the Society for Computer Applications in Radiology. 2012;25(1):14–24. doi: 10.1007/s10278-011-9422-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fenstermacher DA, Wenham RM, Rollison DE, Dalton WS. Implementing personalized medicine in a cancer center. Cancer J. 2011;17(6):528–536. doi: 10.1097/PPO.0b013e318238216e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, Bloomfield CD, Lander ES. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286(5439):531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 81.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B (Methodological) 1995:289–300. [Google Scholar]
- 82.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Storey JD. The positive false discovery rate: A Bayesian interpretation and the q-value. Annals of Statistics. 2003:2013–2035. [Google Scholar]
- 84.Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19(3):368–375. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- 85.Jain A, Dubes R. Algorithms that cluster data. Prentice Hall; Englewood Cliffs, NJ: 1988. [Google Scholar]
- 86.Wilkinson L, Friendly M. The history of the cluster heat map. The American Statistician. 2009;63(2):179–184. [Google Scholar]
- 87.Eschrich S, Yang I, Bloom G, Kwong KY, Boulware D, Cantor A, Coppola D, Kruhoffer M, Aaltonen L, Orntoft TF, Quackenbush J, Yeatman TJ. Molecular staging for survival prediction of colorectal cancer patients. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2005;23(15):3526–3535. doi: 10.1200/JCO.2005.00.695. [DOI] [PubMed] [Google Scholar]
- 88.Shedden K, Taylor JM, Enkemann SA, Tsao MS, Yeatman TJ, Gerald WL, Eschrich S, Jurisica I, Giordano TJ, Misek DE, Chang AC, Zhu CQ, Strumpf D, Hanash S, Shepherd FA, Ding K, Seymour L, Naoki K, Pennell N, Weir B, Verhaak R, Ladd-Acosta C, Golub T, Gruidl M, Sharma A, Szoke J, Zakowski M, Rusch V, Kris M, Viale A, Motoi N, Travis W, Conley B, Seshan VE, Meyerson M, Kuick R, Dobbin KK, Lively T, Jacobson JW, Beer DG. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nature medicine. 2008;14(8):822–827. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jolliffe IT. Principal component analysis. Springer; New York: 2002. p. xxix.p. 487. [Google Scholar]
- 90.Wold S, Ruhe A, Wold H, Dunn W., III The collinearity problem in linear regression. The partial least squares (PLS) approach to generalized inverses. SIAM Journal on Scientific and Statistical Computing. 1984;5:735. [Google Scholar]
- 91.Dupuy A, Simon RM. Critical review of published microarray studies for cancer outcome and guidelines on statistical analysis and reporting. Journal of the National Cancer Institute. 2007;99(2):147–157. doi: 10.1093/jnci/djk018. [DOI] [PubMed] [Google Scholar]
- 92.Simon RM, Paik S, Hayes DF. Use of archived specimens in evaluation of prognostic and predictive biomarkers. Journal of the National Cancer Institute. 2009;101(21):1446–1452. doi: 10.1093/jnci/djp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Efron B. Bootstrap methods: another look at the jackknife. The annals of Statistics. 1979;7(1):1–26. [Google Scholar]
- 94.Efron B. Estimating the error rate of a prediction rule: improvement on cross-validation. Journal of the American Statistical Association. 1983:316–331. [Google Scholar]
- 95.Dougherty ER. Small sample issues for microarray-based classification. Comparative and Functional Genomics. 2001;2(1):28–34. doi: 10.1002/cfg.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kim S, Dougherty ER, Barrera J, Chen Y, Bittner ML, Trent JM. Strong feature sets from small samples. Journal of Computational biology. 2002;9(1):127–146. doi: 10.1089/10665270252833226. [DOI] [PubMed] [Google Scholar]
- 97.Braga-Neto U, Hashimoto R, Dougherty ER, Nguyen DV, Carroll RJ. Is cross-validation better than resubstitution for ranking genes? Bioinformatics. 2004;20(2):253–258. doi: 10.1093/bioinformatics/btg399. [DOI] [PubMed] [Google Scholar]