Abstract
Specimens of neotropical Anopheles (Nyssorhynchus) were collected and identified morphologically. We amplified three genes for phylogenetic analysis–the single copy nuclear white and CAD genes, and the COI barcode region. Since we had multiple specimens for most species we were able to test how well the single or combined genes were able to corroborate morphologically defined species by placing the species into exclusive groups. We found that single genes, including the COI barcode region, were poor at confirming species, but that the three genes combined were able to do so much better. This has implications for species identification, species delimitation, and species discovery, and we caution that single genes are not enough. Higher level groupings were partially resolved with some well-supported groupings, whereas others were found to be either polyphyletic or paraphyletic. There were examples of known groups, such as the Myzorhynchella Section, which were poorly supported with single genes but were well supported with combined genes. From this we can infer that more sequence data will be needed in order to show more higher-level groupings with good support. We got unambiguously good support (0.94–1.0 Bayesian posterior probability) from all DNA-based analyses for a grouping of An. dunhami with An. nuneztovari and An. goeldii, and because of this and because of morphological similarities we propose that An. dunhami be included in the Nuneztovari Complex. We obtained phylogenetic corroboration for new species which had been recognised by morphological differences; these will need to be formally described and named.
Introduction
Malaria is among the world’s most important infectious diseases. There were approximately 216 million cases of malaria reported in 2011 and an estimated 655,000 deaths in 2010 [1]. The disease is a major obstacle to social and economic development in affected countries [2], and although considerable funding has helped decrease the incidence of malaria by an estimated 17% since 2000, the failure to maintain effective malaria control strategies can lead to resurgence in historically endemic regions [1], [3]. Climate change [4] and deforestation [5] are also likely to play an important role in the appearance of malaria in non-endemic and newly inhabited regions, respectively. Effective approaches to species identification are essential in vector incrimination, the assessment of malaria risk and development of malaria control strategies [6]. The focus of such work centres on the genus Anopheles, which contains all known vectors of malaria, with many forming cryptic species complexes [7]. Such species are morphologically indistinguishable and for many of the most important vectors of malaria a molecular approach is the only effective tool for resolving species and species relationships [8].
There are nine dominant Anopheles vector species in the Americas, three species belong to the subgenus Anopheles and six to the Nyssorhynchus [9]. Among species of the subgenus Nyssorhynchus, Anopheles darlingi is the primary vector in areas of the Amazon Region [9], [10]. In addition, An. albimanus, species of the An. albitarsis Complex, An. aquasalis, An. marajoara, and An. nuneztovari are also dominant vectors of human Plasmodium [9]. Other species of the Nyssorhynchus may be secondary, local vectors, or were found naturally infected with Plasmodium sporozoites, for example, An. benarrochi [11], An. rangeli [12], [13], An. oswaldoi [12], [14], An. oswaldoi B [13], An. strodei [15], An. rondoni [16], An. trinkae [12], and An. triannulatus [17].
The Anophelinae subfamily includes 465 formally recognized species and 50 unnamed members of species complexes [7], which are subdivided into three genera – Anopheles, Bironella, and Chagasia. Species of the genus Anopheles are subdivided into seven subgenera: Anopheles, Baimaia, Cellia, Kerteszia, Lophopodomyia, Nyssorhynchus, and Stethomyia. Worldwide, the primary vectors of human malaria parasites belong to the subgenera Anopheles, Cellia and Nyssorhynchus [18]. Most phylogenetic and phylogeographic studies regarding Anophelinae involved medically important species or species groups. The most comprehensive phylogenetic study, in terms of the number of species sampled, is that of Sallum et al. (2000) [19], which employed morphological characters. Results of phylogenetic analysis, using either morphological characters [19] or sequences of mitochondrial and nuclear genes [20] corroborated the monophyly of Anophelinae and of subgenera Cellia, Kerteszia and Nyssorhynchus.
The subgenus Nyssorhynchus has a Neotropical distribution, with An. albimanus extending to the southern Neartic Region. The Nyssorhynchus includes 39 formally named species [21] with a current listing of 44 [22], which are subdivided between the Albimanus Section (Faran 1980) [23], Argyritarsis Section (Linthicum 1988) [24], and Myzorhynchella Section (Galvão 1941) [25]. In the Argyritarsis and Albimanus Sections there are species that were not formally validated and therefore were designated by letters added to the name of the taxon morphologically more similar. This is so in the case of An. nuneztovari which includes chromosomal forms A, B, and C [26]–[28]. Using ITS2 sequence data, Sierra et al. (2004) [29], showed that An. nuneztovari B and C are conspecific, and Calado et al. (2008) [30] demonstrated that at least part of that which was identified as An. nuneztovari A may include An. goeldii, and so this species was resurrected from the synonymy. The Strodei Complex was named by Faran (1980) [23] and includes An. strodei, An. benarrochi and An. rondoni. Anopheles strodei contained five species in synonymy, which were described by Unti [31], [32] using characters either of the eggs or larvae. Recently, Sallum et al. [33] resurrected An. albertoi and An. arthuri from synonymy with An. strodei. The number of recognized Nyssorhynchus species has increased over time, and it is expected that this trend will continue. It has been proposed that An. triannulatus [34], An. konderi [35], [36], and An. oswaldoi [14], [36] may be more than one species under the same name. There are complexes of morphologically similar species in the Argyritarsis Section that are being revealed by genetic and phylogenetic studies using DNA sequences of nuclear and mitochondrial genes, for example five named species and four un-named lineages defined by the COI gene in [37]. This is also so in the An. albitarsis species complex which so far consists of six species [38], [39]. Additionally, the once monospecific Pictipennis Group now includes An. atacamensis, which was recently described from specimens collected in Atacama Desert, Chile. The Myzorhynchella Section consisted until recently of only four species; however it now includes An. antunesi, An. guarani, An. lutzii, An. nigritarsis, An. pristinus and An. parvus. An. guarani was revalidated by Nagaki et al. [40] and An. pristinus described by Nagaki and Sallum (in Nagaki et al. [41]).
Phylogenetic relationships within the subgenus Nyssorhynchus were investigated by Bourke et al. (2010) [42] employing DNA sequence data from the single copy nuclear white gene and the ND6 mitochondrial gene. Generally, results of phylogenetic analyses provided support for some lineages, although the topologies for the two genes disagreed somewhat. Bayesian topology of the combined ND6 and white datasets supported the Myzorhynchella Section as a natural group. Unfortunately, it was not clear if either the Argyritarsis Section or the Albimanus Section was monophyletic, and relationships within these Sections could not be fully assessed because of lack of deep resolution. Furthermore, because of the small species sample size it was not possible to assess monophyly of most groups, subgroups and complexes as defined by Faran (1980) [23] for the Albimanus Section and by Linthicum (1988) [24] for the Argyritarsis Section. However, the topology from the combined dataset confirmed monophyly of the subgroups Oswaldoi and Triannulatus of the Albimanus Section. Regarding the Argyritarsis Section, the An. albitarsis Complex was confirmed as a monophyletic group, and An. lanei as a distinct group, sister to the clade formed by species of the Albimanus and Argyritarsis Sections, without assignment to any Nyssorhynchus section. Inconsistencies and also lack of basal resolution obtained by Bourke et al. (2010) [42] may have been caused by both limited taxa sampling and poor phylogenetic information contained in the two genes employed in the analyses.
Several genes have been found to be useful in Anopheles systematics. The 5′ region of the COI gene has long been used as a DNA barcode for species identification, and also for barcoding animal life [43], [44]. The barcode has been shown to be successful for mosquito identification in Canada [45], and to both corroborate known lineages and define new lineages within the An. albitarsis Complex [37]. Besansky and Fahey [46] used the nuclear white gene to confirm the monophyly of the subfamily Anophelinae, and tribes Sabethini, Culicini and Aedini. More recently, arginine kinase, CAD, catalase, enolase, hunchback, and white nuclear protein-coding genes were used to assess genus level relationships within Culicidae and to infer divergence time for major lineages, strongly supporting the monophyly of Anophelinae and the tribes Aedini and Sabethini [47]. A larger amount of sequence data would allow broader and deeper phylogenetics, such as for example the study by Regier and Zwick [48], who used 62 protein-coding nuclear genes, using only non-synonymous change, to recover a robust higher-level phylogeny of Arthropoda.
Here we used the COI barcode region, and the single copy nuclear white and CAD protein-coding genes to confirm known morphologically defined species, and to estimate phylogenetic relationships and assess species complexes within the subgenus Nyssorhynchus. We found that species confirmation was poor using any of the three genes alone, but was good using concatenated genes. Higher level groupings were partially resolved with some well-supported groupings, whereas others were found to be either polyphyletic or paraphyletic. We propose that An. dunhami be included in the Nuneztovari Complex. We obtained phylogenetic corroboration for new species which had been recognised by morphological differences.
Results and Discussion
Species Identification and DNA-based Species Confirmation
The species and specimens used in this study are listed in Table S1. The species included many of those in the catalog by Harbach [21], and also some undefined morpho-species. These undefined taxa were identified using both male genitalia and fourth-instar larva characteristics, but since they could be differentiated from their morphologically closest relative they were marked as sensu lato; further studies will be necessary to name and validate these taxa. These included taxa in the Oswaldoi Subgroup which were preliminarily identified as An. konderi s.l. [35], [36] and An. oswaldoi s.l. [36], and also An. argyritarsis s.l. The specimens had been identified as above, and also when possible from the scanning electron microscope of the egg; the latter was used to aid identification of species of the Strodei Subgroup and Myzorhynchella Section.
Three genes were used for the molecular analysis – the single copy nuclear white and CAD genes, and the barcode region of the mitochondrial COI gene. The aligned concatenated genes are characterized in Table 1. Model-based phylogenetic analysis was done with two different Bayesian phylogenetic programs, MrBayes v 3.1.2 [49], and p4 [50], and for each program multiple separate “runs” were done. We have as usual pooled the results from the runs and made consensus trees from the pooled samples, but also here we have kept the results of runs separate to better show the spread that support values can take (Figures S1A–E and S2, and Tables S2, S3, and S4). Additionally, pairwise Kimura 2-parameter (K2P) distances were measured among sequences of the COI barcode region, and also of the three concatenated gene regions, and these were used to make neighbour-joining (NJ) trees (Figures S1F and S1G).
Table 1. Characterization of the alignment of the three gene regions.
| gene | codon position | nChar | nTax | unique sequences | constant | variable | parsimony informative |
| concat | 2298 | 144 | 143 | 1428 | 870 | 703 | |
| white | 750 | 137 | 116 | 510 | 240 | 174 | |
| first | 250 | 89 | 204 | 46 | 25 | ||
| second | 250 | 49 | 233 | 17 | 5 | ||
| third | 250 | 114 | 73 | 177 | 139 | ||
| CAD | 846 | 129 | 120 | 440 | 406 | 332 | |
| first | 282 | 108 | 196 | 86 | 53 | ||
| second | 282 | 102 | 224 | 58 | 33 | ||
| third | 282 | 118 | 20 | 262 | 245 | ||
| COI | 702 | 144 | 131 | 478 | 224 | 194 | |
| first | 234 | 103 | 200 | 34 | 26 | ||
| second | 234 | 47 | 233 | 1 | 0 | ||
| third | 234 | 130 | 45 | 189 | 168 |
The outgroup in this analysis are the taxa An. (Kerteszia) cruzii, An. (Anopheles) intermedius, and An. (Stethomyia) kompi. A species of the subgenus Kerteszia was chosen because Kerteszia is a sister-group to Nyssorhynchus; species of the subgenera Anopheles and Stethomyia are somewhat more distant to Nyssorhynchus [19], [20]. Bayesian analysis with all three genes at the DNA level including all three outgroup taxa did not show any support for the outgroup (Figure S1A, Table S2B); it was only when the translations were used that support for the outgroup was seen. Even then, support for a separate outgroup was uneven, ranging from 0.0 to 0.99 Bayesian posterior probability (BPP) in different MCMC runs (Figure S2A, Table S3C). We interpreted this as indicative of unknown biases in the outgroup DNA sequences that were at least partially ameliorated in the translations. Inspection identified An. kompi as a difficult taxon, and it was removed from subsequent DNA-based analyses. When it was removed then the remaining two outgroup taxa generally grouped together, although not with the COI gene, and not fully with the CAD gene (Table S2B).
There were only three of the ingroup species that were represented by a single specimen; all other species in the ingroup were represented by up to ten specimens (Table S1). This allowed us to test how well the DNA-based species reflected the morphological species. We did this by asking whether in a DNA-based phylogenetic tree the specimens formed an exclusive monophyletic clade. Here we are asking whether a single gene contains sufficient phylogenetic information to place a specimen in a species, or whether this placement requires more sequence data.
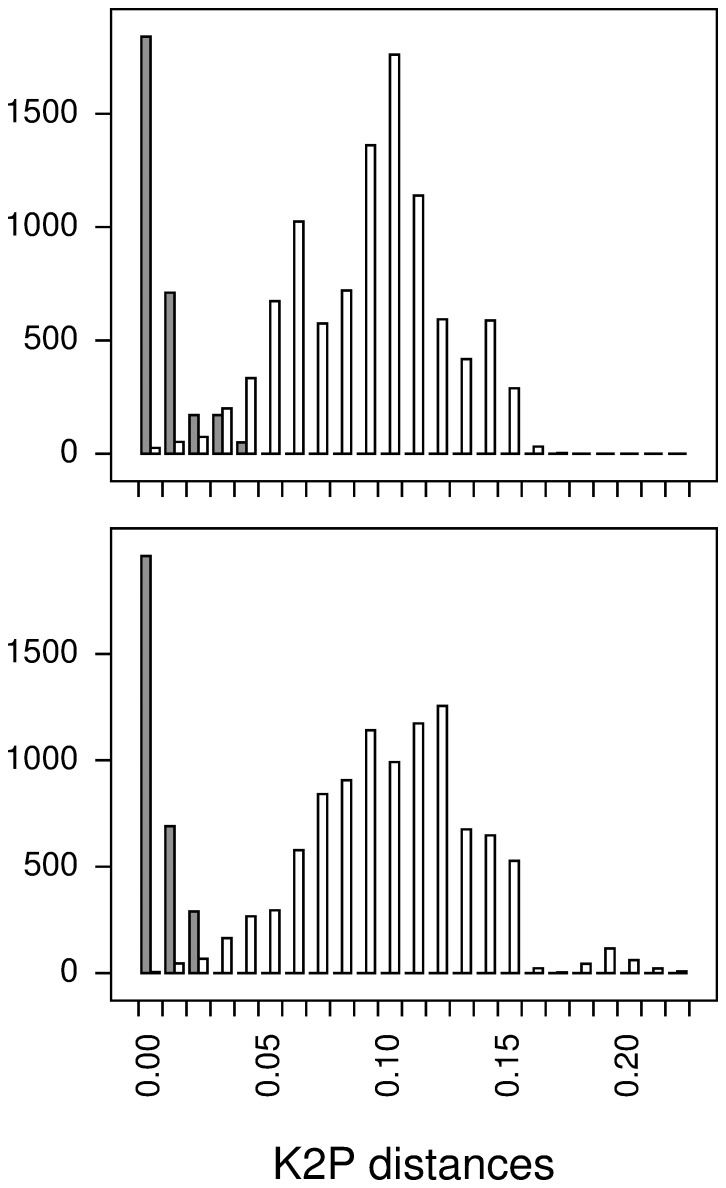
We obtained pairwise Kimura 2-parameter (K2P) distances from the COI barcode region, and from the three gene regions concatenated together, shown in Figure 1. For the COI barcode region, the intraspecific distances ranged from 0.0 to 0.049, while the interspecific distances ranged from 0.0015 to 0.17. For the three concatenated gene regions, the intraspecific distances ranged from 0.0 to 0.029 and the interspecific distances ranged from 0.006 to 0.225. Both the COI barcode region and the three gene region concatenation show considerable overlap in the intra- and interspecific distances, and neither has a clear barcoding gap between them. We can speculate that specimens contributing the largest intraspecific distances (Table S5) should be candidates for being considered separate species; these are discussed below. Specimen pairs with interspecific distances of less than 2% for the COI barcode region were always such that both members of the pair were from within the Oswaldoi Subgroup, within the Nuneztovari Complex, or within the Strodei Subgroup.
Figure 1. Intra- and interspecific K2P distances.
The top panel shows distances between COI barcode regions, and the bottom panel shows distances between specimens using all three gene regions. Dark bars show intraspecific distances and light bars show interspecific distances. The counts of intraspecific distances have been scaled tenfold for clarity.
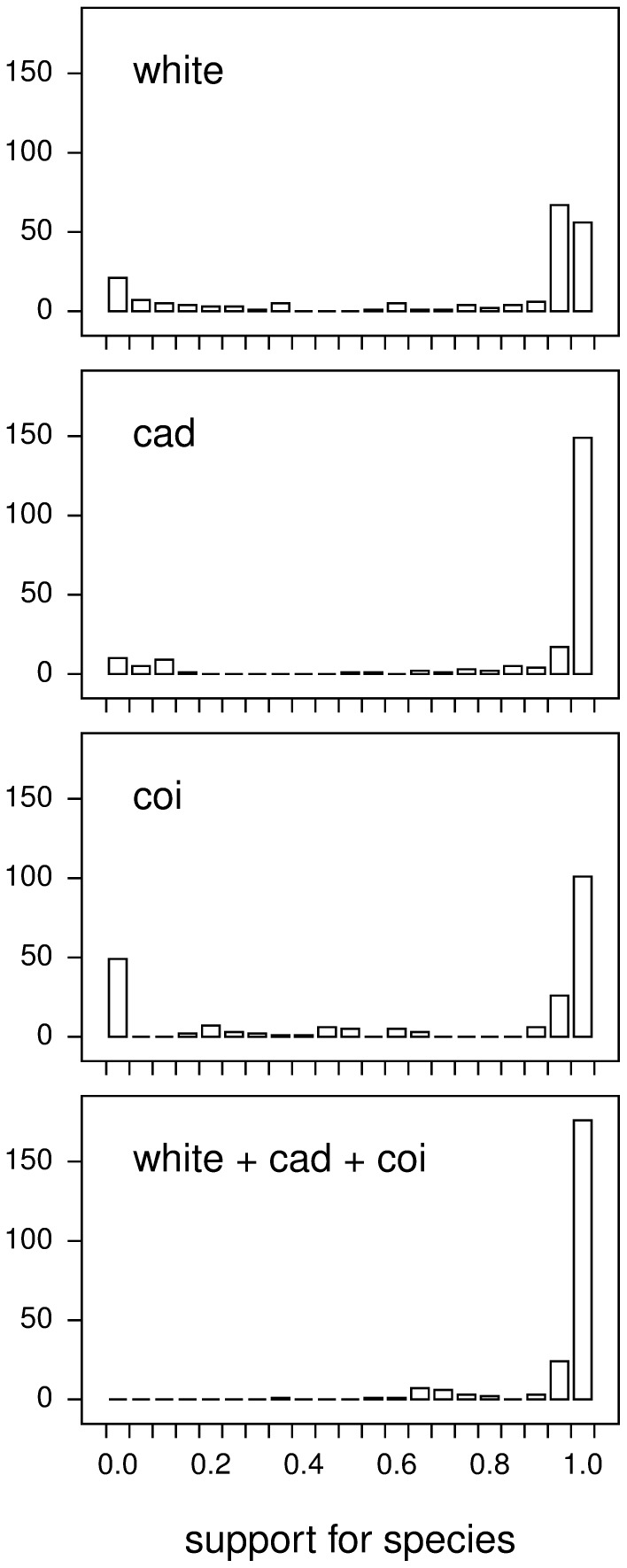
In the Bayesian analyses we can ask how much clade support is given when we use a single gene (one of white, CAD, or COI) compared to the support for species when we use these three genes together. For those species represented by two or more specimens, we obtained support values for monophyly of the specimens for a morphologically defined species (support values were obtained from the MCMC posterior samples, as shown in Table S4, and plotted in Figure 2). The results show that using three genes combined is able to group specimens much better than single genes can do individually. All three single genes, and the COI gene in particular with about 23% of species having less than 10% support, are unable to show support for some species that the combined genes are able to show.
Figure 2. Supports for species using single genes, or using combined data from 3 genes.
Bayesian posterior probabilities of species supports were taken from the posterior distributions (not the consensus trees) of the Bayesian phylogenetic analyses. The rightmost bar shows 100% support. (See Table S4 for specific values.).
These Bayesian analyses were compared to commonly-used K2P/NJ trees (Figure S1F, S1G). Whether these trees showed monophyly for species with more than one specimen is shown in Table 2. When the COI gene was used, 8 of the species were not exclusive in the NJ tree. The COI gene results are similar to the Bayesian analysis using only the COI gene, which had several species with low BPP (Table 2, and Table S6). However, the Bayesian analysis with three genes was much better than the K2P/NJ analysis with the same set of genes (all species in the Bayesian analysis had greater than 0.69 BPP while 6 species were not exclusive in the NJ tree).
Table 2. Species confirmation with COI barcode and with three genes.a .
| species | COI barcode K2P/NJb | COI barcode BPPc | three genes K2P/NJ | three genes BPP |
| An. albertoi | − | 0.00 | + | 1.00 |
| An. albitarsis | + | 0.99 | + | 1.00 |
| An. antunesi | + | 0.49 | − | 1.00 |
| An. argyritarsis | + | 1.00 | + | 1.00 |
| An. arthuri | + | 0.20 | + | 1.00 |
| An. atacamensis | + | 1.00 | − | 1.00 |
| An. benarrochi | + | 1.00 | + | 1.00 |
| An. braziliensis | + | 1.00 | + | 1.00 |
| An. darlingi | + | 1.00 | + | 1.00 |
| An. deaneorum | + | 0.95 | + | 1.00 |
| An. dunhami | + | 0.27 | + | 0.99 |
| An. evansae | − | 0.00 | + | 1.00 |
| An. galvaoi | + | 1.00 | + | 1.00 |
| An. goeldii | − | 0.00 | − | 0.72 |
| An. guarani | + | 1.00 | − | 1.00 |
| An. konderi s.s. | − | 0.99 | + | 1.00 |
| An. konderi A | + | 0.50 | + | 0.99 |
| An. lanei | + | n.a.d | + | 1.00 |
| An. lutzii s.s. | + | 1.00 | + | 1.00 |
| An. lutzii A | + | 0.03 | + | 1.00 |
| An. lutzii B | − | 1.00 | − | 0.69 |
| An. marajoara | + | 0.94 | + | 1.00 |
| An. nuneztovari | − | 0.00 | + | 0.99 |
| An. oswaldoi s.s. | − | 0.00 | − | 0.77 |
| An. oswaldoi A | + | 1.00 | + | 1.00 |
| An. parvus | + | 1.00 | + | 1.00 |
| An. pristinus | + | 1.00 | + | 1.00 |
| An. rangeli | + | 1.00 | + | 1.00 |
| An. rondoni | + | 0.66 | + | 1.00 |
| An. strodei | − | 0.00 | + | 0.86 |
| An. strodei CPform | + | 1.00 | + | 1.00 |
| An. triannulatus | + | 1.00 | + | 1.00 |
DNA-based confirmation of those morphologically-defined species with more than one specimen, using the COI barcode region only or with three genes concatenated.
K2P distances were used to make a NJ tree. “+” if all the specimens formed an exclusive group, or “−” otherwise.
Bayesian posterior probability, averaged from Table S6.
Not applicable; the two COI sequences from An. lanei were identical, and so were homogenized for the Bayesian analysis. This can be taken as BPP of 1.00.
Species Groups
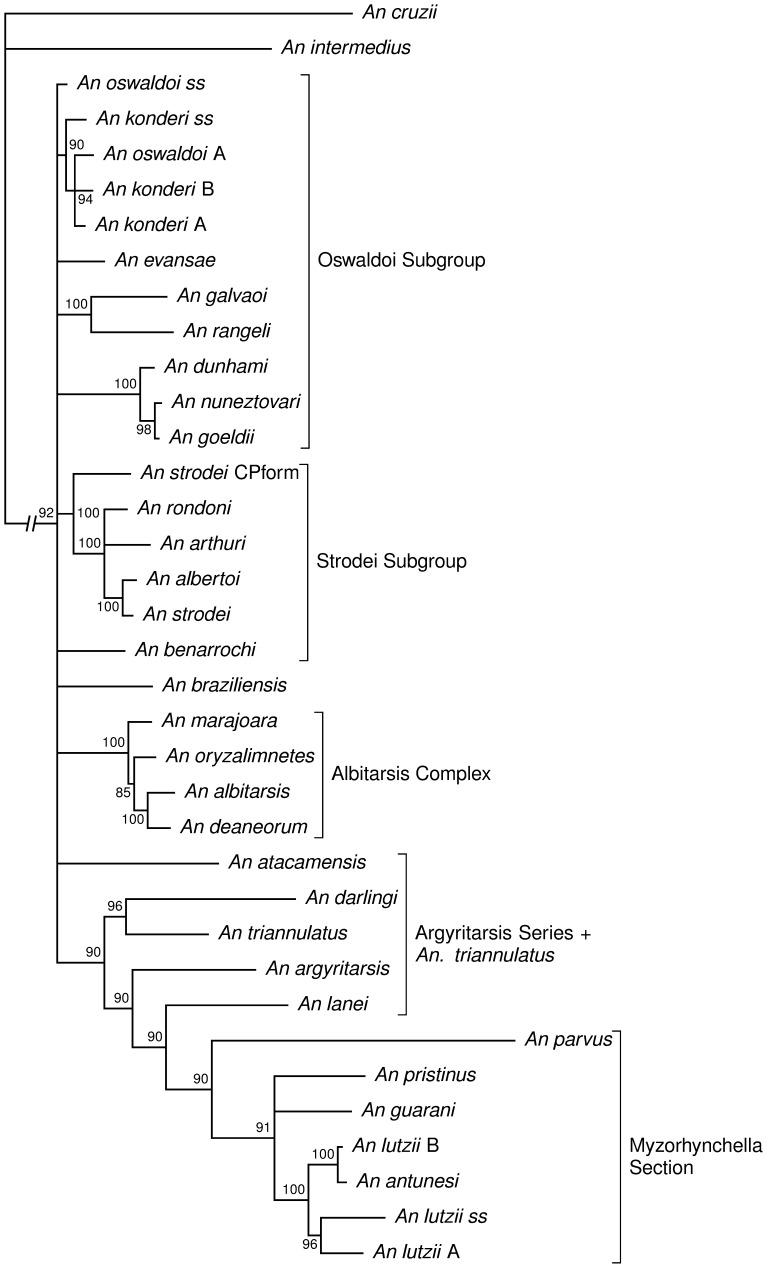
Using all three genes, the consensus tree shown in Figure 3 was obtained. The species groups supported by the analyses in this study are described with reference to Harbach [21]. An. triannulatus and An. braziliensis were found to be more or less unstable taxa in the analyses described below; for example the position of An. triannulatus in Figure 3 is certainly questionable. Agreement between our phylogenetic analysis and the current classification hierarchy was uneven. Of the three Sections that we sampled from (the Albimanus, Myzorhynchella, and Argyritarsis Sections), only the Myzorhynchella Section had support as a clade; it had support from analysis of translations (0.77–0.97 BPP), and support from combined genes (0.70–1.0 BPP), but not from individual genes (0.0–0.12 BPP, with a single 0.8 BPP; Tables S2 and S3). Patterns seen within these Sections are described here (note that we only sampled from the Oswaldoi Series from the Albimanus Section).
Figure 3. Consensus tree from a Bayesian phylogenetic analysis using all three gene regions.
This is a species-level depiction of Figure S1B; see that figure for analysis details. Internal nodes show support as posterior probability, expressed as percent.
Oswaldoi series
The Oswaldoi Series is composed of the Oswaldoi Group and the Triannulatus Group, and in turn the Oswaldoi Group is composed of the Oswaldoi Subgroup and the Strodei Subgroup. There is little support for the Oswaldoi Subgroup; there is some, but uneven, support for this grouping from the combined DNA analysis (0.0–0.9 BPP), but not from any of the three genes individually (0.0–0.14 BPP, Table S2). Within the Oswaldoi Subgroup, there is support for various groupings from the DNA analyses (the translations do not resolve within this Complex). Harbach [21] cites the Nuneztovari Complex within the Oswaldoi Subgroup, composed of An. goeldii and An. nuneztovari [30]. We got support from most analyses for a grouping of An. goeldii and An. nuneztovari consistent with the Nuneztovari Complex (0.91–1.0 BPP using all three gene regions), but we got consistent support from all DNA analyses for this grouping including An. dunhami (0.94–1.0), and we suggest that An. dunhami should be placed in this Complex. All three species can be easily recognized as morphologically similar species by characteristics of the male genitalia [51]. For example, the ventral claspette has a truncate, wide apex, and the basal lobules are moderately expanded laterally with spicules along basal margin; these spicules are moderately long and evenly distributed over the basal surface and radiate in different directions, and the ventral and lateral surfaces of the ventral claspette are covered with short setae. Considering Faran’s 1980 identification key for the female and male genitalia and fourth-instar larva, the key characters used to identify An. nuneztovari can be applied as characteristics for the Nuneztovari Complex, since it is likely that the specimens examined by him were a mix of all three species [23]. Also within the Oswaldoi Subgroup we see some support for a grouping of (An. konderi s.s., (An. konderi A, An. konderi B, An. oswaldoi A)) (0.81–1.0 BPP using all three gene regions), discussed below, and for An. galvaoi with An. rangeli (1.0 BPP using all three gene regions) (Figure 3).
None of the analyses show support for the Strodei Subgroup. The combined DNA analysis supports the remaining members of the Strodei Subgroup if An. benarrochi is excluded (1.0 BPP). Most analyses, both DNA and translation, support a grouping of An. albertoi, An. strodei, An. arthuri, and An. rondoni (1.0 BPP using all three gene regions), with support from combined DNA analysis for An. albertoi with An. strodei (1.0 BPP using all three gene regions). There is some, but uneven, support for the Oswaldoi Group (that is the Oswaldoi Subgroup with the Strodei Subgroup) from combined DNA analysis (0.0–1.0 BPP) and from COI by itself (0.66–0.78 BPP).
The position of An. triannulatus is not resolved with these data. In particular, there is no support for the Oswaldoi Series (including An. triannulatus, excluding An. braziliensis). In combined DNA analyses An. triannulatus goes as sister group to An. darlingi, while in translations there is some support for An. triannulatus with An. rangeli (0.26–0.51 BPP).
Argyritarsis section
There is wide and fairly good support for the Albitarsis Group/Complex, from both DNA and translation analysis (1.0 BPP using all three genes, 0.56–0.99 BPP using translations, but 1.0 BPP when using translations with no outgroup; Tables S2 and S3). However, there is no support for the Albitarsis Series, which here would mean the Albitarsis Group with An. braziliensis, the latter found to be unstable in our analyses. These and other taxa of the Argyritarsis Section are generally near each other in the tree as a grade, but with no support as a group.
Myzorhynchella section
There is good support in all analyses, including analyses of the individual genes, for a crown group composed of all taxa sampled from this section except An. parvus (0.72–1.0 BPP using all three gene regions; Table S2). Support for the Myzorhynchella Section, with An. parvus basal, is generally good (0.70–1.0 using all three gene regions), but oddly somewhat uneven when the outgroup taxon An. kompi is included. None of the three genes separately support the Myzorhynchella Section, but together they do support it (with the white gene and the CAD gene the placement of An. parvus is ambiguous, however the COI gene places An. parvus in the outgroup).
Evidence for New Species and a New Subgenus
Identification of the species of the subgenus Nyssorhynchus is complicated by the presence of polymorphisms and overlap of morphological characters used in identification keys. Consequently DNA sequence evidence has been used to aid in recognition of species. Marrelli et al. (1999) [14] used nucleotide sequences of the second internal transcribed spacer (ITS2) of ribosomal DNA to revise the taxonomic status of An. oswaldoi, suggesting that there were at least four sibling species under the name An. oswaldoi. Among those species was An. konderi, which was subsequently redescribed and removed from the synonymy with An. oswaldoi by Flores-Mendoza et al. (2004) [52]. Unfortunately Marrelli et al. (1999) [14] did not include an An. konderi specimen from the type locality (Coari, State of Amazonas, Brazil), and so the molecular identity of An. konderi remains undefined. In a recent review, Marrelli et al. (2006) [53] showed that one of the sequences generated by Marrelli et al. (1999) [14], belonged to an individual of An. evansae which had been mistakenly identified as An. oswaldoi. More recently, Sallum et al. (2008) [36] noted that specimens collected in Acre and preliminarily identified as An. konderi could be differentiated by morphological characteristics of the male genitalia from other An. konderi specimens and also from that defined by Flores-Mendoza et al. (2004) [52]. In addition, differences in the sequences of the ITS2 rDNA corroborated a morphological hypothesis that suggested the presence of undescribed species that could be confused with An. konderi when using only female characters. Continuing these studies, Motoki et al. (2011) [35] increased An. konderi sampling, including representatives of the states of Paraná, Rondônia, Acre and Amapá, and sequences from the ITS2 rDNA, COI barcode region of mtDNA and the single copy nuclear white gene. As expected, An. konderi was confirmed as a species complex. The originality of results obtained by Motoki et al. (2011) [35] was that the name An. konderi includes at least three species that can be separated by both morphological traits of aedeagus of the male genitalia and by DNA sequence. However, further studies will be necessary to establish accurate comparisons and validate the genetic lineages observed by Sallum et al. (2008) [36] and Motoki et al. (2011) [35]. Our results in this study described above show support for a grouping of (An. konderi A, (An. konderi s. s., An. konderi B, An. oswaldoi A)) (0.90 BPP; Figure 3 and Figure S1B, see also Table S1 for specimen identity).
The cluster formed by An. argyritarsis and An. argyritarsis s.l. (MG25_4) seems to be composed of two distinct species; individuals from CE20, CE17 and MG04 represent An. argyritarsis, whereas specimen MG25_4 is likely of a distinct species (Figure S1B). Morphological characteristics of the fourth-instar larva, mainly seta 3-T of the metathorax and seta 1-I of the abdominal segment I, allow separation of An. argyritarsis s.l. MG25_4 from the remaining An. argyritarsis (CE20, CE17 and MG04). Additionally, female abdominal scales key out the individual as An. sawyeri, whereas larval setal characteristics do not key out the specimen either as An. argyritarsis or An. sawyeri. Considering morphological differences observed in the specimen MG25_4 regarding to both An. argyritarsis and An. sawyeri, and to avoid premature species definition, that individual was assigned to the morphologically closest taxon, until male and female linked to larval and pupal exuviae are obtained in field collections, and DNA sequence can be associated to specimen identified with accuracy.
Until recently the Myzorhynchella Section consisted of four named species; currently it is composed of six valid taxa – An. antunesi, An. guarani, An. lutzii, An nigritarsis, An. pristinus and An. parvus. Anopheles guarani was resurrected from synonymy of An. lutzii by Nagaki et al. (2011) [40], and An. pristinus was described by Nagaki and Sallum (in Nagaki et al. 2010, [41]). Recently, Bourke et al. (2011) [54] suggested that individuals identified as An. lutziiA325 and An. lutziiB369 may represent two unnamed species. Additional specimens obtained in the same geographical region of An. lutziiA325 and An. lutziiB369 confirmed that they are two unnamed taxa that can be misidentified as An. lutzii if the identification is based only on external characters of the female. One cluster of individuals (including RS19 and RS33, tentatively labelled as An. lutzii A in Figure 3 ) seems to be more closely related to An. lutzii s.s. than to any other species of the Myzorhynchella Section. The second cluster, including RS16a and RS16b, together with B369, tentatively labelled An. lutzii B in Figure 3, is sister to An. antunesi (see Table S1 for specimen identity). The COI barcode distances between B369 and the two RS16 specimens is 4.9% (Table S5), suggesting that B369 is a species separate from the other two. The hypothesis of three unnamed taxa is corroborated by morphological traits of the male genitalia and fourth-instar larva. The new species will be formally described and named in a further study by one of the authors.
Other specimens with large intraspecific distances (Table S5) which would be candidates for being considered separate species include An_parvus_MG07_9_1, An_triannulatus_ES03_03_01, An_oswaldoi_SP22_9, and An_evansae_SP12_44.
If we look at pairs with the largest (>16%) K2P distances we can see that one member of the pair was always a specimen of An. parvus. These distances are larger than seen in intra-subgenus distances where one member of the pair was from the outgroup taxa An. cruzii or An. intermedius; these large genetic distances suggest that An. parvus should be considered a distinct subgenus. This is corroborated by morphological differences. Differences in the eggs as seen with SEM [55] and in male genitalia traits can easily separate An. parvus from all other species on the subgenus Nyssorhynchus. For example, the presence of spicules in the mesal margin of the ventral lobule of the ventral claspette is a characteristic that is not observed in any species of the Myzorhynchella section, and the parabasal lobule is curved, not straight as in species of the Nyssorhynchus. An. parvus was described as a species of the genus Myzorhynchella (currently a section), which has Anopheles niger as the type species. The latter species is in synonymy with An. lutzii, and consequently the name Myzorhynchella cannot be used for An. parvus if it is determined to be a species of a distinct subgenus. Whether Myzorhynchella is a section of the subgenus Nyssorhynchus or a subgenus of the genus Anopheles is an open question to be answered in further studies.
Based on phylogenetic analysis (Figure S1), it was speculated that An. nuneztovari specimen RO2_13 was misidentified, and indeed this specimen seems to be a representative of An. goeldii that was misidentified by morphological traits as An. nuneztovari. Consequently, both species may be sympatric in Rondônia state, Brazil, extending geographical distribution of An. goeldii to the western Amazon River basin.
Conclusions
-
A single gene region is often not sufficient to place specimens in morphologically defined species. The COI barcode region is for species identification, and has been used to not only corroborate morphologically defined species but also to define new species in the Anopheles albitarsis Complex [37]. We have tested the ability of COI, white, and CAD to group specimens known to be in the same species together, and have shown that individual genes often are unable to do this, while the concatenated genes are much better at delineating species.
The method of analysis affects the veracity of the observation above. Using the simple K2P/NJ method there was only a small improvement with an increase in the amount of sequence data. However, with a Bayesian analysis and better fitting models there was a greater improvement (Table 2).
-
We need more sequence data in order to show higher level groupings. It may be that the noise and uncertainty that we see is due to systematic error, in which case we need better methods or models, or to poor sampling, in which case we need more sequence data. If few data give us poor resolution or poor support, but having more sequence data allows us to obtain results that are consistent with well-established relationships, that argues that we need primarily more sequence data.
We have examples from this study that support this. For example, we have some support for Oswaldoi Subgroup from the combined DNA analysis, but not from any of the three genes individually. Another example is that none of the three genes separately support the Myzorhynchella Section, but together they do support it. However, our study leaves too many unanswered questions regarding the phylogenetic relationships in the subgenus Nyssorhynchus, even with three genes. It is an easy prediction that more sequence information will help in this regard, and an obvious target would be to get mitochondrial genomes, which have helped resolve similar problems elsewhere [56]–[59].
Translations can be useful for deeper phylogenetic distinctions. Analysis of the translations, that is analysis at the amino acid level, can give results that make more sense than the DNA analysis; for example the position of An. kompi in the outgroup was seen with translations but not with DNA. However, the translation sequences are more similar to each other, as is expected within a subgenus, and so there is poor resolution between closely related taxa. What we see in this case using translations is an unresolved comb phylogeny.
We propose that An. dunhami should be part of the Nuneztovari Complex.
-
We have some phylogenetic evidence for new species, corroborated by morphological evidence.
An. argyritarsis s.l. may represent a new species, morphologically similar to both An. argyritarsis and An. sawyeri.
Specimens of the tentatively named An. oswaldoi A from Pará state and Acre (AC18) are likely representatives of an undescribed species that is morphologically similar to An. oswaldoi.
An. konderi from Amapa state and AC18_16 from Acre state seem to belong to two species closely related to An. konderi that have not yet been formally named. Specimens of An. konderi from Rondônia and Paraná state may represent An. konderi s.s. because the type locality is Coari, Amazonas state, situated south of the Amazon River basin. Additionally, morphological comparison of male genitalia of one specimen from Coari collected by Flores-Mendoza in 1998 shows that representatives from Rondônia and Paraná states are morphologically more similar to that of Coari than to those from individuals from Amapa and Acre states.
Tentatively-named An luzii A, B, and B369 appear to be separate species.
There are several specimens with large intraspecific K2P distances (>3%) that are candidates for being considered separate species. These include An_parvus_MG07_9_1, An_triannulatus_ES03_03_01, An_oswaldoi_SP22_9, and An_evansae_SP12_44.
Based on morphological differences and very large (>16%) K2P distances, An. parvus should possibly be considered a separate subgenus of Anopheles.
Materials and Methods
Ethics Statement
All necessary permits were obtained for the described field studies. Collections were made under permanent permit number 16938-1 from Instituto Brasileiro do Meio Ambiente e dos Recursos Naturais Renováveis (IBAMA) to Maria Anice M. Sallum. Specific permission was not required for these locations as permission to collect was granted under the permanent permit. The collection locations were not privately owned or protected in any way. The field studies did not involve protected or endangered species.
Taxon Sampling
The species sampled for this study and the sources of specimens are listed in Table S6. Larvae and pupae were either collected from field habitats or obtained from link-reared offspring of blood fed females collected in the field. Both larvae and pupae were maintained in the laboratory to obtain adult males and females associated with larval and pupal exuviae. Freshly emerged mosquitoes were quickly anesthetized with ethyl acetate vapors, and either kept separate in minute plastic vials in silica gel or individually frozen at −80°C. Species identification was based on either adult male genitalia or fourth-instar larval characteristics. For few taxa, identification was also based on the external morphology of the eggs observed in a Jeol JSM-6460 scanning electron microscope (SEM) (Jeol Ltd., Akishima, Japan) as reported by Sallum et al. (2010) [33] and Nagaki et al. (2010) [41].
DNA extraction. DNA was extracted from whole mosquitos, following the insect DNA extraction protocol provided by the QIAgen DNeasy Blood and Tissue Kit (QIAgen Ltd., Crawley, UK).
DNA Amplification and Sequencing
COI mtDNA
The primers LCO1490 (5′ GGT CAA CAA ATC ATA AAG ATA TTG G 3′) and HCO2198 (5′ TAA ACT TCA GGG TGA CCA AAA AAT CA 3′) (Table S7) were used to amplify about 650 base pairs of the mitochondrial cytochrome subunit I (COI mtDNA). Each PCR reaction contained 1 µl template DNA (about 1/200th to 1/1000th of the amount extracted from a single specimen); 20 mM Tris-HCl, pH 8.4; 50 mM KCl; 1.5 mM MgCl2; 5 picomoles of each primer; 200 µM each dNTPs; and 2.5 U Taq polymerase; and the remaining volume of ultra pure water up to 25 µl. PCR amplification protocol consisted of a 3-min denaturation at 94°C and 35 cycles at 94°C, 55° and 72°C for 1 minute each, followed by a 7 minute extension at 72°C.
Nuclear white gene
Amplification of about 700 base pairs of the nuclear single copy white gene was obtained using the primers WZ2E and WZ11X (Table S7). Each PCR reaction contained 1–2 µl template DNA (about 1/100th to 1/200th of the amount extracted from a single specimen); 20 mM Tris-HCl, pH 8.4; 50 mM KCl; 1.5 mM MgCl2; 2.5 µl DMSO; 100 picomoles of each primer; 200 µM each dNTPs; 2.5 U Taq polymerase; and the remaining volume of ultra pure water up to 25 µl. PCR amplification protocol consisted of a 3-minute denaturation at 94°C, 35 cycles at 94°C for 30 sec, 50°C for 1 min and 72°C for 2 min each, followed by a 7-min extension at 72°C. PCR amplicons obtained from six females were purified using PEG precipitation (20% polyethylene glycol 8000/2.5 M NaCl) and cloned into pGem-T Easy Vector (Promega, Madison, WI, USA). Two to four positive clones were sequenced.
Nuclear CAD gene
Amplification of about 648–895 base pairs of the nuclear single copy CAD gene was obtained using the primers listed in Table S7. Each PCR reaction contained 2 to 4 µl template DNA (about 1/50th to 1/100th of the amount extracted from a single specimen); 20 mM Tris-HCl, pH 8.4; 50 mM KCl; 1.5 to 2.4 mM MgCl2; 2.5 µl DMSO; 100 picomoles of each primer; 200 µM each dNTPs; 2.5 U Taq polymerase; and the remaining volume of ultra pure water up to 25 to 50 µl. PCR amplification protocol consisted of a 2 to 4-min denaturation at 94°C, 35 cycles at 94°C for 20 to 30 sec, 50°C to 57°C for 30 sec and 72°C for 1 min each, followed by a 7-min extension at 72°C. For the PCR employing CAD338F_M13 and CAD680R_M13, the protocol consisted of a 3-min denaturation at 94°C, 35 cycles at 94°C for 1 min, 56°C for 1 min and 72°C for 1 min each, followed by a 7-min extension at 72°C. PCR amplicons obtained from 13 individuals were purified using PEG precipitation (20% polyethylene glycol 8000/2.5 M NaCl) and cloned into pGem-T Easy Vector (Promega, Madison, WI, USA). One to four positive clones were sequenced.
PCR products of white, CAD, and COI genes were electrophoresed in 1% TAE agarose gels stained with GelRedTM Nucleic Acid Gel Stain (Biotium Inc., Hayward, USA). All sequencing reactions were carried out in both directions using ABI Big Dye Terminator Kit v.3.1 (PE Applied Biosystems, Warrington, England). For COI we employed the same primers used for PCR, whereas for the white and CAD genes, primers were variable and are listed in Table S7. Sequencing reaction consisted of 0.5 µl of Big Dye Terminator Ready Reaction Mix; 3 µl of 5X sequence dilution-buffer [5 mM MgCl2, 200 mM Tris-HCl, pH 9.0]; 3.6 picomoles of R or F primer; 10 ng of PEG purified PCR product, ultra pure water up to 10 µl. Sequencing reactions were purified in Sephadex G50 columns (GE Healthcare). Sequences were analyzed on an ABI Prism 3130 – Avant Genetic Analyzer (Applied Biosystems, Foster City, CA, U.S.A.), and edited using Sequencher for Windows version 4.9 (Gene Codes Corporation, Ann Arbor, USA).
DNA vouchers
Template DNA from this study is retained at −70°C in the Faculdade de Saúde Pública (FSP-USP), São Paulo, Brazil, for future reference (DNA reference numbers are in Table S6). Immature and male genitalia slide of the specimens used for DNA extraction are deposited in the FSP-USP collection.
Sequence Data Preparation and Analysis
Among the 144 specimens, all had a COI sequence, 137 had a white sequence, and 129 had a CAD sequence (Table S8). DNA sequences were translated using Genewise v 2.2.0 [60], [61], using closely related protein guide sequences from GenBank and appropriate genetic codes. The resulting amino acid sequences were aligned with Clustalo (v 1.0.2, [62]). Inspection of the alignments showed that the white gene and the CAD gene had unreliable alignment sites, which were masked out by hand. The codons used to make the translations were back-aligned to the protein alignments, and correspondingly masked. There was an intron in the white gene (although not all sequences had that intron); separate alignment of the intron DNA sequence with Muscle [63] did not give a satisfactory alignment, so the intron was not used further.
Analysis using K2P/NJ
Pairwise Kimura 2-parameter (K2P) [64] pairwise distances were calculated from the aligned COI barcode region and the three aligned concatenated genes, both without An. kompi, using PAUP* [65]. Trees were made from these distances with the neighbour joining (NJ) algorithm [66], again as implemented in PAUP*.
Bayesian phylogenetic analysis
The aligned concatenated genes are characterized in Table 1. Notice that there are no informative sites in the COI second codon position; it was not used in the Bayesian analyses.
Preliminary analysis showed that partitioning by gene and by codon was better than partitioning by either alone. Models were chosen for the eight partitions in isolation, aided by Modeltest v 3.7 [67]. Modeltest was done on the separate partitions after removing blank sequences, homogenizing duplicate sequences, and then using only likelihood informative sites. The models suggested are tabulated in Table S9, together with the AIC weight. Often the suggested model was not implemented in the software used to do the analysis (MrBayes or p4), and so the model actually used is also shown, together with its AIC weight. Often the AIC weights were low (generally 0.2 or less), indicating ambiguity in model choice.
Only the 81 topology informative sites of the 766 sites in the concatenated translation were used. In these sequences 15 of the 144 sequences had more than 40 gaps in the alignment of 81 sites, and those gappy sequences were removed as being too short. However, one of those short sequences was An. kompi, one of the outgroup sequences, and this sequence was retained for some analyses even though it was short. Finally, duplicate sequences were homogenized, leaving 106 sequences (including An. kompi). Model choice was aided by Prottest v3.0 [68], which indicated that the JTT+G model was best by the AIC criterion, with an AIC Weight of 0.999.
Supporting Information
Phylogenetic trees from DNA. Consensus trees of Bayesian and K2P/NJ analysis of individual and concatenated gene regions. See the caption in the Figure for details of analysis. (A) Analysis of DNA of all three gene regions, including An. kompi in the the outgroup. (B) Analysis of all three gene regions, not including An. kompi in the the outgroup. (C) Analysis of the white gene only. (D) Analysis of the CAD gene only. (E) Analysis of the COI barcode region only. (F) K2P/NJ tree of the COI barcode region, not including An. kompi in the outgroup. (G) K2P/NJ tree of the DNA from all three sequences, not including An. kompi in the outgroup.
(PDF)
Phylogenetic trees from protein translations. Consensus trees of Bayesian analysis of translations of concatenated gene regions. See the caption in the Figure for details of the analysis. (A) Analysis of protein translations of all three gene regions, including An. kompi in the the outgroup. (B) Analysis of protein translations of all three gene regions, not including An. kompi in the the outgroup. (C) Analysis of protein translations of all three gene regions, not including the outgroup.
(PDF)
Species and specimens. A table showing the specimens used in this study, and the morphologically-based species identification.
(PDF)
Support for higher-level groupings from Bayesian analyses of DNA sequences. Support for species is shown in Table S6. Support is shown as Bayesian posterior probability as taken from the posterior distribution; that is from the sampled trees, not from a consensus tree.
(PDF)
Support for species and higher-order groupings from gene translations. Support is shown as Bayesian posterior probability as taken from the posterior distribution; that is from the sampled trees, not from a consensus tree.
(PDF)
Support for species from three concatenated gene regions and from single gene regions only. Support is shown as Bayesian posterior probability as taken from the posterior distribution; that is from the sampled trees, not from a consensus tree. Support for higher-level groupings is shown in Table S4.
(PDF)
Intraspecifc K2P distances greater than 3%: Possible candidates for new species. One of the pair would be a possible candidate for placement in another species.
(PDF)
Specimen collection locations and details.
(XLS)
DNA amplification primers.
(DOC)
Specimens and sequences used. A table showing which sequences were used in the analysis, and what name was given to those sequences.
(PDF)
Model choice. Tables showing the models suggested by Modeltest and the models that were used in the model-based DNA analyses. The Akaike weights were often low, showing ambiguity in the model choice.
(PDF)
Acknowledgments
Thanks go to the two reviewers, who provided suggestions and comments that greatly improved the first draft of the manuscript.
Funding Statement
This investigation was financially supported by the Fundação de Amparo a Pesquisa do Estado de São Paulo (FAPESP) (Grants no. 2011/20397-7; 2005/53973-0; http://www.fapesp.br/), and Conselho Nacional de Desenvolvimento Técnico e Científico (CNPq BPP no. 301666/2011-3 to MAMS; http://www.cnpq.br/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.World Health Organization (2011) World Malaria Report 2011. Geneva: World Health Organization.
- 2. Sachs J, Malaney P (2002) The economic and social burden of malaria. Nature 415: 680–685. [DOI] [PubMed] [Google Scholar]
- 3. Cohen J, Smith D, Cotter C, Ward A, Yamey G, et al. (2012) Malaria resurgence: a systematic review and assessment of its causes. Malaria J 11: 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chaves L, Koenraadt C (2010) Climate change and highland malaria: fresh air for a hot debate. Quart Rev Biol 85: 27–55. [DOI] [PubMed] [Google Scholar]
- 5. Vittor A, Pan W, Gilman R, Tielsch J, Glass G, et al. (2009) Linking deforestation to malaria in the amazon: characterization of the breeding habitat of the principal malaria vector, Anopheles darlingi . Am J Trop Med Hyg 81: 5–12. [PMC free article] [PubMed] [Google Scholar]
- 6. Zapata M, Cienfuegos A, Quirós O, Quiñones M, Luckhart S, et al. (2007) Discrimination of seven anopheles species from san pedro de urabá, antioquia, colombia, by polymerase chain reaction–restriction fragment length polymorphism analysis of its sequences. Am J Trop Med Hyg 77: 67–72. [PubMed] [Google Scholar]
- 7.Harbach RE (1818) Genus Anopheles Meigen, 1818. Available: http://mosquito-taxonomic-inventory.info/genus-emanophelesem-meigen-1818-0. Accessed 2012 Jul 19.
- 8. White B, Collins F, Besansky N (2011) Evolution of anopheles gambiae in relation to humans and malaria. Ann Rev Ecol Evol Syst 42: 111–132. [Google Scholar]
- 9. Sinka ME, Rubio-Palis Y, Manguin S, Patil AP, Temperley WH, et al. (2010) The dominant Anopheles vectors of human malaria in the Americas: occurrence data, distribution maps and bionomic précis. Parasit Vectors 3: 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Oliveira-Ferreira J, Lacerda MVG, Brasil P, Ladislau JLB, Tauil PL, et al. (2010) Malaria in Brazil: an overview. Malar J 9: 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Flores-Mendoza C, Fernández R, Escobedo-Vargas K, Vela-Perez Q, Schoeler G (2004) Natural Plasmodium infections in Anopheles darlingi and Anopheles benarrochi (Diptera: Culicidae) from eastern Peru. J Med Entomol 41: 489–494. [DOI] [PubMed] [Google Scholar]
- 12. Hayes J, Calderon G, Falcon R, Zambrano V (1987) Newly incriminated anopheline vectors of human malaria parasites in Junin Department, Peru. J Am Mosq Control Assoc 3: 418–422. [PubMed] [Google Scholar]
- 13. Quiñones ML, Ruiz F, Calle DA, Harbach RE, Erazo HF, et al. (2006) Incrimination of Anopheles (Nyssorhynchus) rangeli and An. (nys.) oswaldoi as natural vectors of Plasmodium vivax in Southern Colombia. Mem Inst Oswaldo Cruz 101: 617–23. [DOI] [PubMed] [Google Scholar]
- 14. Marrelli MT, Malafronte RS, Flores-Mendoza C, Lourenço-de-Oliveira R, Kloetzel JK, et al. (1999) Sequence analysis of the second internal transcribed spacer of ribosomal dna in Anopheles oswaldoi (Diptera: Culicidae). J Med Entomol 36: 679–84. [DOI] [PubMed] [Google Scholar]
- 15. de Oliveira-Ferreira J, Lourenço-de-Oliveira R, Teva A, Deane LM, Daniel-Ribeiro CT (1990) Natural malaria infections in anophelines in Rondonia State, Brazilian Amazon. Am J Trop Med Hyg 43: 6–10. [PubMed] [Google Scholar]
- 16. da Rocha J, de Oliveira S, Póvoa M, Moreira L, Krettli A (2008) Malaria vectors in areas of Plasmodium falciparum epidemic transmission in the Amazon region, Brazil. Am J Trop Med Hygiene 78: 872–877. [PubMed] [Google Scholar]
- 17. de Arruda M, Carvalho M, Nussenzweig R, Maracic M, Ferreira A, et al. (1986) Potential vectors of malaria and their different susceptibility to Plasmodium falciparum and Plasmodium vivax in northern Brazil identified by immunoassay. Am J Trop Med Hygiene 35: 873–881. [DOI] [PubMed] [Google Scholar]
- 18. Sinka M, Bangs M, Manguin S, Rubio-Palis Y, Chareonviriyaphap T, et al. (2012) A global map of dominant malaria vectors. Parasit Vectors 5: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sallum MAM, Schultz TR, Wilkerson RC (2000) Phylogeny of Anophelinae (Diptera Culicidae) based on morphological characters. Annals Entomol Soc Am 93: 745–775. [Google Scholar]
- 20. Sallum MAM, Schultz TR, Foster PG, Aronstein K, Wirtz RA, et al. (2002) Phylogeny of Anophelinae (Diptera: Culicidae) based on nuclear ribosomal and mitochondrial DNA sequences. Syst Entomol 27: 361–382. [Google Scholar]
- 21.Harbach RE (1902) Subgenus Nyssorhynchus Blanchard, 1902. Available: http://mosquito-taxonomic-inventory.info/subgenus-nyssorhynchus-blanchard-1902. Accessed 2012 Jul 19.
- 22.Harbach RE. Subgenus Nyssorhynchus. Available: http://mosquito-taxonomic-inventory.info/subgenus-ltemgtnyssorhynch. Accessed 2012 Jul 19.
- 23.Faran M (1980) Mosquito studies (Diptera, Culicidae). XXXIV. A revision of the Albimanus Section of the subgenus Nyssorhynchus of Anopheles. Contrib Am Entomol Inst 15.
- 24.Linthicum K (1988) A revision of the Argyritarsis section of the subgenus Nyssorhynchus of Anopheles (Diptera: Culicidae). Technical report, DTIC Document.
- 25. Galvão A (1941) Contribuição ao conhecimento das esp´ecies de myzorhynchella (Diptera: Culicidae). Arch Zool S Paulo 2: 505–576. [Google Scholar]
- 26. Kitzmiller J, Kreutzer R, Tallaferro E (1973) Chromosomal differences in populations of Anopheles nuneztovari . Bull World Health Org 48: 435. [PMC free article] [PubMed] [Google Scholar]
- 27. Conn J (1990) A genetic study of the malaria vector Anopheles nuneztovari from western Venezuela. J Am Mosq Contr Assoc 6: 400–405. [PubMed] [Google Scholar]
- 28. Conn J, Puertas YR, Seawright JA (1993) A new cytotype of Anopheles nuneztovari from western Venezuela and Colombia. J Am Mosq Control Assoc 9: 294–301. [PubMed] [Google Scholar]
- 29. Sierra DM, Velez ID, Linton YM (2004) Malaria vector Anopheles (Nyssorhynchus) nuneztovari comprises one genetic species in Colombia based on homogeneity of nuclear ITS2 rDNA. J Med Entomol 41: 302–7. [DOI] [PubMed] [Google Scholar]
- 30. Calado DC, Foster PG, Bergo ES, Santos CLSd, Galardo AKR, et al. (2008) Resurrection of Anopheles goeldii from synonymy with Anopheles nuneztovari (Diptera, Culicidae) and a new record for Anopheles dunhami in the Brazilian Amazon. Mem Inst Oswaldo Cruz 103: 791–9. [DOI] [PubMed] [Google Scholar]
- 31. Unti O (1940) Anofelinos do Vale do Paraiba. Nota iii. Biologia do Anofeles [sic] (Nyssorhynchus) strodei Rooth, 1926 com a descrição d’uma variedade nova. Anofeles [sic] (Nyssorhynchus) strodei ramosi var. Ann Paulist Med Cir 40: 489–505. [Google Scholar]
- 32. Unti O (1941) Anofelinos do vale do Rio Paraiba, Anopheles (Nyssorhynchus) strodei Root 1926, com a descrição de três variedades novas. São Paulo Serv Profil Mal Trab 33: 3–18. [Google Scholar]
- 33. Sallum MAM, Foster PG, Dos Santos CLS, Flores DC, Motoki MT, et al. (2010) Resurrection of two species from synonymy of Anopheles (Nyssorhynchus) strodei Root, and characterization of a distinct morphological form from the Strodei complex (Diptera: Culicidae). J Med Entomol 47: 504–26. [DOI] [PubMed] [Google Scholar]
- 34. Silva-do Nascimento TF, Lourenço-de Oliveira R (2007) Diverse population dynamics of three Anopheles species belonging to the triannulatus complex (diptera: Culicidae). Mem Inst Oswaldo Cruz 102: 975–82. [DOI] [PubMed] [Google Scholar]
- 35. Motoki MT, Bourke BP, Bergo ES, Da Silva AM, Sallum MAM (2011) Systematic notes of Anopheles konderi and its first record in Parańa State, Brazil. J Am Mosq Control Assoc 27: 191–200. [DOI] [PubMed] [Google Scholar]
- 36. Sallum MAM, Marrelli MT, Nagaki SS, Laporta GZ, Dos Santos CLS (2008) Insight into Anopheles (Nyssorhynchus) (Diptera: Culicidae) species from Brazil. J Med Entomol 45: 970–81. [DOI] [PubMed] [Google Scholar]
- 37. Ruiz-Lopez F, Wilkerson RC, Conn JE, McKeon SN, Levin DM, et al. (2012) DNA barcoding reveals both known and novel taxa in the Albitarsis group (Anopheles: Nyssorhynchus) of neotropical malaria vectors. Parasit Vectors 5: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Brochero HHL, Li C, Wilkerson RC (2007) A newly recognized species in the Anopheles (Nyssorhynchus) albitarsis complex (Diptera: Culicidae) from Puerto Carreno, Colombia. Am J Trop Med Hyg 76: 1113–7. [PubMed] [Google Scholar]
- 39. Motoki MT, Wilkerson RC, Sallum MAM (2009) The Anopheles albitarsis complex with the recognition of Anopheles oryzalimnetes Wilkerson and Motoki, n. sp. and Anopheles janconnae Wilkerson and Sallum, n. sp. (Diptera: Culicidae). Mem Inst Oswaldo Cruz 104: 823–50. [DOI] [PubMed] [Google Scholar]
- 40. Nagaki SS, Da Silva AM, Sallum MAM (2011) Redescription of Anopheles (Nyssorhynchus) lutzii, and resurrection of Anopheles guarani from synonymy with An. lutzii (Diptera: Culicidae). Ann Entomol Soc Am 104: 374–388. [Google Scholar]
- 41. Nagaki SS, Motta MdA, Sallum MAM (2010) Redescription of Anopheles (Nyssorhynchus) antunesi Galvão & Amaral and description of a new species of the Myzorhynchella Section (Diptera: Culicidae) from Serra da Mantiqueira, Brazil. Mem Inst Oswaldo Cruz 105: 278–85. [DOI] [PubMed] [Google Scholar]
- 42. Bourke BP, Foster PG, Bergo ES, Calado DC, Sallum MAM (2010) Phylogenetic relationships among species of Anopheles (Nyssorhynchus) (Diptera, Culicidae) based on nuclear and mitochondrial gene sequences. Acta Trop 114: 88–96. [DOI] [PubMed] [Google Scholar]
- 43. Hebert PDN, Cywinska A, Ball SL, de WaardJR (2003) Biological identifications through DNA barcodes. Proc Roy Soc Ser B: Biol Sci 270: 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hebert P, Ratnasingham S, deWaard J (2003) Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc Roy Soc Ser B: Biol Sci 270: S96–S99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cywinska A, Hunter FF, Hebert P (2006) Identifying Canadian mosquito species through DNA barcodes. Med Veterinary Entomol 20: 413–424. [DOI] [PubMed] [Google Scholar]
- 46. Besansky NJ, Fahey GT (1997) Utility of the white gene in estimating phylogenetic relationships among mosquitoes (Diptera: Culicidae). Mol Biol Evol 14: 442–54. [DOI] [PubMed] [Google Scholar]
- 47. Reidenbach KR, Cook S, Bertone MA, Harbach RE, Wiegmann BM, et al. (2009) Phylogenetic analysis and temporal diversification of mosquitoes (Diptera: Culicidae) based on nuclear genes and morphology. BMC Evol Biol 9: 298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Regier JC, Zwick A (2011) Sources of signal in 62 protein-coding nuclear genes for higher-level phylogenetics of arthropods. PLoS One 6: e23408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–4. [DOI] [PubMed] [Google Scholar]
- 50. Foster PG (2004) Modeling compositional heterogeneity. Syst Biol 53: 485–95. [DOI] [PubMed] [Google Scholar]
- 51. Causey O (1945) Description of Anopheles (Nyssorhynchus) dunhami, a new species from the Upper Amazon Basin. J Natl Malar Soc 4: 231–234. [Google Scholar]
- 52. Flores-Mendoza C, Peyton E, Wilkerson R, Oliveira R (2004) Anopheles (Nyssorhynchus) konderi Galvão and Damasceno: neotype designation and resurrection from synonymy with Anopheles (Nyssorhynchus) oswaldoi (Peryassú)(Diptera: Culicidae). Proc Entom Soc Washington 106: 118–132. [Google Scholar]
- 53. Marrelli MT, Sallum MAM, Marinotti O (2006) The second internal transcribed spacer of nuclear ribosomal DNA as a tool for Latin American anopheline taxonomy–a critical review. Mem Inst Oswaldo Cruz 101: 817–32. [DOI] [PubMed] [Google Scholar]
- 54. Bourke BP, Nagaki SS, Bergo ES, Cardoso JdC, Sallum MAM (2011) Molecular phylogeny of the Myzorhynchella section of Anopheles (Nyssorhynchus) (Diptera: Culicidae): genetic support for recently described and resurrected species. Mem Inst Oswaldo Cruz 106: 705–15. [DOI] [PubMed] [Google Scholar]
- 55. Forattini O, Sallum M, Bergo E, Flores D (1998) Ultrastructure of eggs of anopheles rondoni, anopheles lutzii, and anopheles parvus. three species of the subgenus nyssorynchus. J Am Mosq Control Assoc 14: 256–265. [PubMed] [Google Scholar]
- 56. Cameron S, Lambkin C, Barker S, Whiting M (2007) A mitochondrial genome phylogeny of Diptera: whole genome sequence data accurately resolve relationships over broad timescales with high precision. Syst Entomol 32: 40–59. [Google Scholar]
- 57. Fenn JD, Song H, Cameron SL, Whiting MF (2008) A preliminary mitochondrial genome phylogeny of Orthoptera (Insecta) and approaches to maximizing phylogenetic signal found within mitochondrial genome data. Mol Phylogenet Evol 49: 59–68. [DOI] [PubMed] [Google Scholar]
- 58. Krzywinski J, Li C, Morris M, Conn JE, Lima JB, et al. (2011) Analysis of the evolutionary forces shaping mitochondrial genomes of a Neotropical malaria vector complex. Mol Phylogenet Evol 58: 469–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Talavera G, Vila R (2011) What is the phylogenetic signal limit from mitogenomes? The reconciliation between mitochondrial and nuclear data in the Insecta class phylogeny. BMC Evol Biol 11: 315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Birney E, Durbin R (1997) Dynamite: a flexible code generating language for dynamic programming methods used in sequence comparison. In: Proc. Int. Conf. Intell. Syst. Mol. Biol. volume 5: 56–64. [PubMed] [Google Scholar]
- 61.Birney E, Copley R (2012) Wise2. Available: http://www.ebi.ac.uk/~birney/wise2. Accessed 2012 Dec 21.
- 62.Sievers F, Wilm A, Dineen D, Gibson T, Karplus K, et al.. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Molec syst biol 7. [DOI] [PMC free article] [PubMed]
- 63. Edgar R (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic acids research 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Molec Evol 16: 111–120. [DOI] [PubMed] [Google Scholar]
- 65.Swofford DL (2002) PAUP*. Phylogenetic analysis using parsimony (*and other methods), Version 4. Sinauer Associates, Sunderland, Massachusetts.
- 66. Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol biol and evol 4: 406–425. [DOI] [PubMed] [Google Scholar]
- 67. Posada D, Crandall K (1998) Modeltest: testing the model of DNA substitution. Bioinformatics 14: 817–818. [DOI] [PubMed] [Google Scholar]
- 68. Darriba D, Taboada G, Doallo R, Posada D (2011) ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27: 1164–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic trees from DNA. Consensus trees of Bayesian and K2P/NJ analysis of individual and concatenated gene regions. See the caption in the Figure for details of analysis. (A) Analysis of DNA of all three gene regions, including An. kompi in the the outgroup. (B) Analysis of all three gene regions, not including An. kompi in the the outgroup. (C) Analysis of the white gene only. (D) Analysis of the CAD gene only. (E) Analysis of the COI barcode region only. (F) K2P/NJ tree of the COI barcode region, not including An. kompi in the outgroup. (G) K2P/NJ tree of the DNA from all three sequences, not including An. kompi in the outgroup.
(PDF)
Phylogenetic trees from protein translations. Consensus trees of Bayesian analysis of translations of concatenated gene regions. See the caption in the Figure for details of the analysis. (A) Analysis of protein translations of all three gene regions, including An. kompi in the the outgroup. (B) Analysis of protein translations of all three gene regions, not including An. kompi in the the outgroup. (C) Analysis of protein translations of all three gene regions, not including the outgroup.
(PDF)
Species and specimens. A table showing the specimens used in this study, and the morphologically-based species identification.
(PDF)
Support for higher-level groupings from Bayesian analyses of DNA sequences. Support for species is shown in Table S6. Support is shown as Bayesian posterior probability as taken from the posterior distribution; that is from the sampled trees, not from a consensus tree.
(PDF)
Support for species and higher-order groupings from gene translations. Support is shown as Bayesian posterior probability as taken from the posterior distribution; that is from the sampled trees, not from a consensus tree.
(PDF)
Support for species from three concatenated gene regions and from single gene regions only. Support is shown as Bayesian posterior probability as taken from the posterior distribution; that is from the sampled trees, not from a consensus tree. Support for higher-level groupings is shown in Table S4.
(PDF)
Intraspecifc K2P distances greater than 3%: Possible candidates for new species. One of the pair would be a possible candidate for placement in another species.
(PDF)
Specimen collection locations and details.
(XLS)
DNA amplification primers.
(DOC)
Specimens and sequences used. A table showing which sequences were used in the analysis, and what name was given to those sequences.
(PDF)
Model choice. Tables showing the models suggested by Modeltest and the models that were used in the model-based DNA analyses. The Akaike weights were often low, showing ambiguity in the model choice.
(PDF)