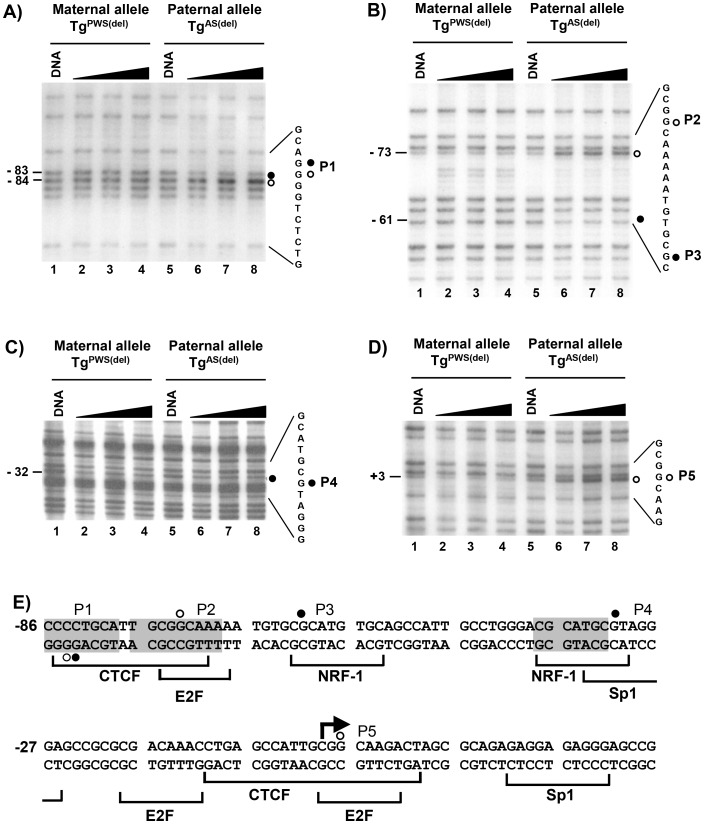
Figure 2. In vivo footprint analysis of the Snrpn promoter region.
LMPCR in vivo footprinting with dimethyl sulfate (DMS) was performed on primary brain cells isolated from TgPWS(del) and TgAS(del) mice carrying a deletion of the entire murine AS/PWS region on either the paternal or maternal chromosomes, respectively [20]. Cells were treated with increasing time of exposure to DMS. “DNA” represents control samples of purified genomic DNA subjected to DMS treatment and LMPCR in parallel with samples treated in vivo with increasing time of exposure to DMS (60, 90, 120 seconds). The LMPCR primer sets detecting each footprint are described in the text. A) – D) Autoradiograms showing in vivo footprints P1 – P5 in the Snrpn promoter region. Filled circles represent footprints detected as guanines protected from DMS reactivity, open circles represent footprints at guanine sites showing enhanced DMS reactivity. Numbers on the left of each sequencing gel denote the location of each footprint relative to the transcription initiation site. The nucleotide sequences to the right of each autoradiogram show the DNA sequence containing and flanking each footprint; the filled/open circles indicate the location of each in vivo footprint within the DNA sequence and correspond to the footprinted sites shown on the left of each autoradiogram. E) Summary of in vivo footprints in the Snprn promoter region of the paternal allele. The footprinted sites detected by LMPCR in vivo footprinting are shown within the nucleotide sequence of the promoter region of the mouse Snprn gene. Open and filled circles denote footprinted sites as described above. At each footprinted position, the footprinted site corresponds to the strand containing the guanine nucleotide. Shaded nucleotide sequences indicate sequences conserved between the human and mouse Snrpn promoters. Brackets below the nucleotide sequence indicate the location and identity of potential transcription factor binding sites. The bent arrow depicts the transcription initiation site; numbering of nucleotides is relative to the transcription initiation site.