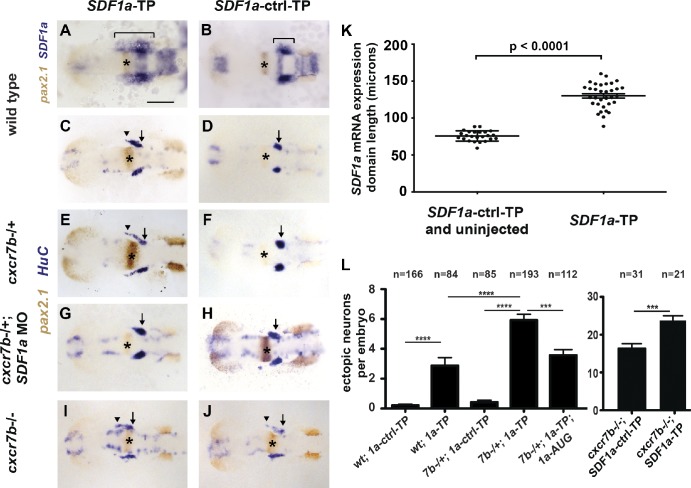
Figure 3.
miR-430 is required for TgSN migration. 10-somite-stage embryos in A and B were stained for with SDF1a (blue) and pax2.1 (brown, MHB marked with asterisks) mRNA. Embryos in C–J were stained for HuC (blue, neurons) and pax2.1 (brown, MHB with asterisks) mRNA. Arrows and arrowheads in C–J denote correctly and incorrectly positioned TgSNs, respectively. Dorsal view, anterior to the left. (A and C) Wild-type embryos injected with SDF1a-TP. (B and D) Wild-type embryos injected with SDF1a-ctrl-TP. (E and F) cxcr7b−/+ embryos injected with SDF1a-TP or SDF1a-ctrl-TP. (G and H) cxcr7b−/+ embryos injected with SDF1a-TP or SDF1a-ctrl-TP and a suboptimal dose of a translation blocking SDF1a morpholino. (I and J) cxcr7b−/− embryos injected with SDF1a-TP or SDF1a-ctrl-TP. (K) Quantification of the SDF1a expression domain in SDF1a-TP embryos (n = 33) compared with controls (n = 8 for SDF1a-ctrl-TP, n = 16 for uninjected wild type). Anterior–posterior lengths in microns correspond to the bracketed region in A and B. Student’s t test: P < 0.0001. (L) Quantification of TgSN positioning defects. Neurons adjacent or anterior to the MHB were defined as ectopic. Student’s t test: ***, P = 0.001; ****, P < 0.0001. See also Table 1. Bar, 100 µm. Data are shown as mean ± SEM (error bars).