Abstract
Predator-prey interactions presumably play major roles in shaping the composition and dynamics of microbial communities. However, little is understood about the population biology of such interactions or how predation-related parameters vary or correlate across prey environments. Myxococcus xanthus is a motile soil bacterium that feeds on a broad range of other soil microbes that vary greatly in the degree to which they support M. xanthus growth. In order to decompose predator-prey interactions at the population level, we quantified five predation-related parameters during M. xanthus growth on nine phylogenetically diverse bacterial prey species. The horizontal expansion rate of swarming predator colonies fueled by prey lawns served as our measure of overall predatory performance, as it incorporates both the searching (motility) and handling (killing and consumption of prey) components of predation. Four other parameters – predator population growth rate, maximum predator yield, maximum prey kill, and overall rate of prey death – were measured from homogeneously mixed predator-prey lawns from which predator populations were not allowed to expand horizontally by swarming motility. All prey species fueled predator population growth. For some prey, predator-specific prey death was detected contemporaneously with predator population growth, whereas killing of other prey species was detected only after cessation of predator growth. All four of the alternative parameters were found to correlate significantly with predator swarm expansion rate to varying degrees, suggesting causal inter-relationships among these diverse predation measures. More broadly, our results highlight the importance of examining multiple parameters for thoroughly understanding the population biology of microbial predation.
Introduction
Animal predators play major roles in regulating ecosystem dynamics [1, 2] both by directly affecting prey populations and by indirectly affecting non-prey species with which prey interact [3]. Because predatory phenotypes and their community-level effects are determined by multiple traits, characterization of those traits can provide important information about how specific features of predation influence community dynamics. Recent expansions of optimal foraging theory have modeled how foraging mode can impact the energetic flux [4] and structure [5] of whole communities. For example, the two spider species Pisaurina mira and Phidippus rimator hunt for Melanopuls femurrubrum grasshoppers by sit-and-wait vs. active searching modes, respectively, and these distinct hunting modes have been shown to differentially impact plant community composition in the grasslands these spiders inhabit [6]. Natural microbial communities also harbor numerous predators [7–9], but little is currently known about their roles in determining the of structure, dynamics and functions of such microbial communities. However, classic experiments in simplified laboratory systems [10, 11], the broad phylogenetic distribution of microbial predators and their prey [12] and the pervasiveness of microbial predators throughout both terrestrial and aquatic habitats [13, 14] all strongly suggest that they play major roles in regulating the dynamics of microbial prey populations [11, 15].
Myxococcus xanthus is a globally distributed soil bacterium that can feed on other microbial cells both individually [16] and in swarming groups that have been likened to wolf packs [17]. Two major aspects of M. xanthus predatory behavior are roughly analogous to the traditional distinction between searching vs. handling components of animal predation [18, 19]. M. xanthus cells “search” for other microbes to prey upon by swarming through the soil matrix powered by two genetically distinct, yet complementary, mechanisms of gliding motility [20, 21]. Upon encounter, M. xanthus “handles” prey cells by secreting molecules that kill and degrade them [22–24] and then consuming their remains as a growth substrate. M. xanthus predation is thought to be a cooperative trait because secretion of predation-associated molecules (e.g. hydrolytic enzymes) might benefit not only secreting cells themselves but neighboring cells as well [17]. M. xanthus can utilize a wide variety of bacteria and fungi as prey, but these vary greatly in the degree to which they support Myxococcus population growth [25, 26] .
The expansion rate of M. xanthus colonies that simultaneously utilize prey as a growth substrate and increase their spatial territory by gliding motility [27] is commonly used as a metric of overall predatory performance on agar plates [28–30]. This performance measure incorporates both the searching and handling components of predation that are traditionally distinguished in foraging theory [31]. For example, Hillesland et al. allowed M. xanthus populations to swarm across plates containing spatially separate patches of prey bacteria and distinguished between the rate of searching for new patches vs. consumption (i.e. handling) of prey within patches [19]. Some studies have also quantified the dynamics or extent of prey death due to M. xanthus predation under conditions of colony expansion [26, 29]. However, it remains unclear which such alternative quantitative parameters are predictive of overall predatory performance.
In this study, we measured both the rate at which M. xanthus populations expand while swarming across clonal lawns of diverse prey species and four additional parameters that were measured while predator populations were increasing in number but not expanding the size of their circumscribed territories. These other four parameters - predator population growth rate, maximum predator yield, rate of prey death, and maximum extent of prey death – are related to prey “handling” [18] because they emerge from direct interaction of predator and prey. These handling-related parameters were analyzed to assess the degree to which they correlate with a standard measure of overall predatory performance (rate of predator swarming expansion across prey lawns) and one another. These analyses allowed us to explore several previously unexamined aspects of microbial predation. For instance, it is unknown whether population growth rate during predation in the absence of group swarming is predictive of overall performance when predators are both increasing numerically and expanding their two-dimensional territory via gliding motility. Additionally, relationships between the dynamics and extent of predator growth and prey death remain obscure.
The rate of M. xanthus swarm expansion on prey lawns emerges from interaction between cell division rate and the dynamics of directional cell motility at a swarm’s leading edge. Leading-edge cells migrate away from the high-density interior of the swarm, where the interior sub-population reaches stationary phase upon depletion of growth substrates extracted from prey. One force that may affect this interaction is any predator Allee effect (i.e. benefit of high density [32, 33]) on prey handling efficiency that may occur at a swarm’s leading edge. It has been hypothesized that M. xanthus predation may be more efficient (i.e. more effectively convert prey biomass into predator biomass) at high predator density than at low density [17]. If this occurs, then growth dynamics at a swarm’s leading edge, where cells foray from high-density cell waves to interact with prey cells, may differ substantially from those in the non-swarming arenas in which we measured population growth rate, predatory yield, prey death rate and extent of prey death. In these arenas, predator and prey cells were dispersed randomly across the entire surface of an agar plate at a ratio of ~1:100,000. Thus, predator cells began growth as isolated individuals or in small cell clumps. Resulting predator microcolonies in this arena then grew together across the entire plate surface and subsequent population growth reflected increasing local density rather than expansion into new territory. Given the differences in the local growth environments of dividing cells in the swarming vs. non-swarming growth arenas examined here, any relationships between parameters across these environments are not obvious a priori.
Material and Methods
Species and culture conditions
Nine phylogenetically diverse species of bacteria used as prey in previous studies of M. xanthus predation [25] were examined here (Table 1), including several species thought to predominantly reside in soil habitats. Prey species were obtained from the Long Term Ecological Research (LTER) collection from Kellogg Biological Station, Hickory Corners, Michigan [34]; the Leibniz Institute German Collection of Microorganisms and Cell Cultures (DSMZ); the American Type Culture Collection (ATCC), and Richard Lenski [35] (Table 1). M. xanthus strain GJV2 was used as the predator and is a spontaneous rifampicin-resistant mutant of strain GJV1, a recent isolate [36] of the commonly studied reference strain DK1622 [37] and is highly proficient at development and motility [38].
TABLE 1.
Prey species.
| Prey species (abbreviation) | Phylum and subdivision | Source [reference] | Strain Reference no. |
|---|---|---|---|
| Arthrobacter globiformis (AG) | Gram positive, High G+C subdivision | LTER [34] | LTER 27 |
| Bacillus bataviensis (BB) | Gram positive, Low G+C subdivision | DSMZ | 15601 |
| Curtobacterium citreum (CC) | Gram positive, High G+C subdivision | LTER [34] | LTER 17 |
| Cytophaga johnsonae (CJ) | Gram positive, Low G+C subdivision | ATCC | 17061 |
| Escherichia coli REL606 (EC) | Gram negative, Gamma subdivision | Richard Lenski [35] | N/A |
| Micrococcus luteus (ML) | Gram positive, Low G+C subdivision | ATCC | 4698 |
| Pseudomonas fluorescens (PF) | Gram negative, Gamma subdivision | LTER [34] | LTER 56 |
| Rhizobium vitris (RV) | Gram negative, Alpha subdivision | DSMZ | 6583 |
| Xanthomonas fragariae (XF) | Gram negative, Gamma subdivision | DSMZ | 3587 |
LTER: Long Term Ecological Research (LTER) site at Kellogg Biological Station, Hickory Corners, Michigan (http://lter.kbs.msu.edu); DSMZ (Leibniz Institute German Collection of Microorganisms and Cell Cultures (www.dsmz.de); ATCC: American Type Culture Collection (www.atcc.org).
To prepare prey suspensions, prey samples were inoculated from frozen stocks into eight ml of fresh R2 broth [39] and grown for 16 hours at 32 °C, 300 rpm. Cells were then centrifuged (4500 g, 15 min.) and resuspended to a standard biovolume concentration of ~109 μm3/ml in liquid TPM buffer [40]. 100 μl of resuspended culture were spread across LB 1.5% agar plates (15 ml) prepared 24 hours prior to inoculation of prey and incubated at 32 °C, 90% rH for 48 h. Cells were then scraped into ten ml of TPM buffer, centrifuged and resuspended in fresh TPM to ~1010 μm3/ml.
Cultures of M. xanthus were first grown on CTT 1.5% agar plates (4–5 days) and then inoculated from the colony edge into CTT broth [37] and grown overnight at 32 °C, 300 rpm. To initiate all assays, log-phase cultures of M. xanthus (~2–3 × 108 cells/ml) were centrifuged (4500 g, 15 min.) and resuspended in liquid TPM buffer to ~5 × 109 cells/ml.
Estimates of M. xanthus growth and prey death
Eight ml aliquots of TPM or CTT 1.5% agar were allowed to solidify in 50 ml flasks two days prior to each experiment and were then kept covered at room temperature. Shortly before inoculation, 5–7 sterile glass beads (3 mm) were placed in the flasks to allow even spread of inocula. To inoculate TPM flasks with predator-prey mixes, 100 μl of a culture containing ~109 μm3 prey cells and ~104 M. xanthus cells (diluted from the 5 × 109 cells/ml suspension) were spread across the agar surface. We used small and large initial predator and prey population sizes, respectively, to allow tracking of substantial predator population growth. As controls, prey-only inocula were spread onto TPM agar and predator-only inocula were spread onto both TPM and CTT agar. Flasks were then left open in laminar flow hood for 15 min., covered and incubated at 32 °C, 90% rH.
Cultures were harvested 2, 12, 24, 36, 48, 60, 84, 108, and 132 h after inoculation to estimate predator and prey population sizes. To harvest, five ml of TPM liquid were added to the flasks and the lawn of cells was separated from the agar surface by repeated pipetting of the TPM liquid. Cell suspensions were then transferred to a 50 ml Falcon tube containing 20–30 sterile glass beads (3 mm) and vortexed vigorously for 90 sec. Suspensions were diluted in TPM liquid and plated either into CTT 0.5% agar containing 10 μg/ml gentamycin sulfate for M. xanthus counts or onto LB 1.5% agar for prey counts. All prey used are sensitive to gentamycin and M. xanthus strain GJV2 is unable to grow on LB medium. Plates were incubated at 32 °C and were counted after two days for prey plates and six days for M. xanthus plates. Predator growth curves and prey death curves in the presence and absence of the predator were then generated from this data.
Swarming rate assays
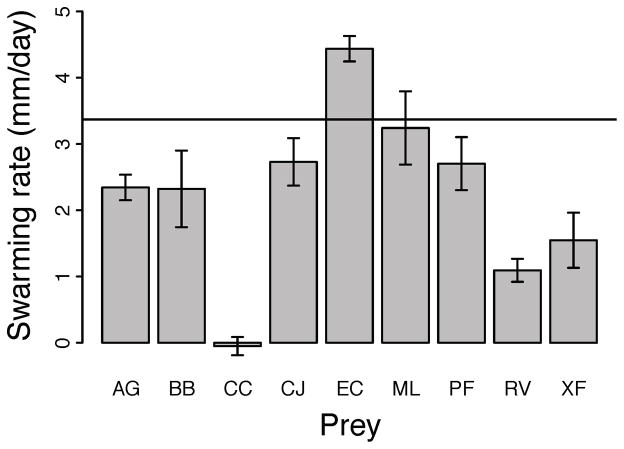
Prey lawns for swarming assays were prepared by spreading 100 μl of prey suspension (~1010 μm3/ml) onto TPM 1.5% agar plates (15 ml TPM, prepared 24 h prior to inoculation). After 30 min. at room temperature, 10 μl of the predator suspension (~5 × 109 cells/ml) were spotted on the plate center and allowed to dry for 60 min. Covered plates were then incubated at 32 °C, 90% rH. Swarm edges were outlined one and five days after inoculation and distances between time-point marks along two perpendicular diameters at random orientation were measured. Prey-specific swarming rates on prey lawns relative to prey-free controls were calculated as the average distance (mm) per day covered by the advancing swarm edge minus the M. xanthus swarming rate on starvation agar controls (Fig. 1). On prey-free TPM control plates, M. xanthus cells undergo starvation and fruiting body formation in the interior of the initially inoculated region, but cells at the perimeter do swarm outward to a limited degree.
Figure 1.
Average M. xanthus swarming rate across prey lawns spread on starvation agar standardized by swarming rate on prey-free starvation plates (see Methods). A value of zero corresponds to equal swarming on prey vs. prey-free starvation agar plates. The solid line indicates the average M. xanthus swarming rate on nutrient rich CTT agar in the absence of prey. Error bars indicate the standard error of the mean.
Parameter estimation
Predator growth rate was calculated as the average slope of ln-transformed population size estimates over the 24–60 h. interval. This period was chosen to factor out any residual growth over the initial 24 h. period that might be due to nutrients acquired by cells during the pre-conditioning phase in CTT liquid or growth on nutrients present at very low concentrations in agar [25]. (M. xanthus populations increased on prey-free TPM plates over the initial 24 h. period and then ceased growing (Fig. 2)). In all prey environments except Curtobacterium citreum (CC) predator growth declined substantially or ceased after the 60 h. time point. To measure overall prey kill rate the average difference between the slopes of ln-transformed estimates of prey population size from 2 h. – 132 h. on TPM vs. TPM+predator plates was calculated. The maximum prey kill was estimated as the maximum difference observed across time points between ln-transformed prey counts in predator-free flasks and counts from flasks containing M. xanthus, standardized by the predator-free counts. Maximum predator yield was calculated as the largest population size reached by M. xanthus over the entire duration of the growth assays (132 hours).
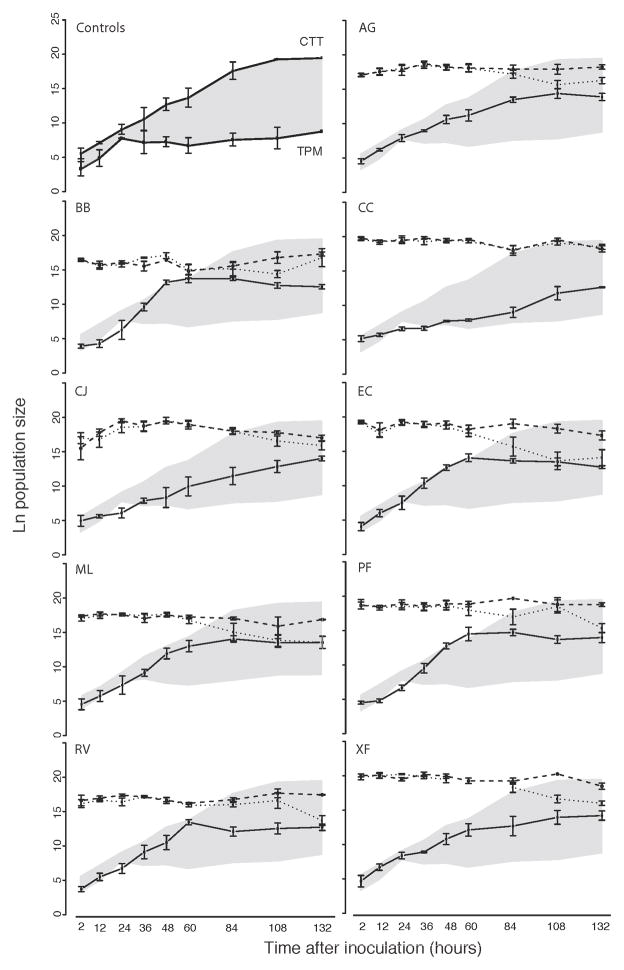
Figure 2.
Population dynamics. Controls (upper left): ln-transformed population size estimates of M. xanthus on starvation agar (bottom line) and nutrient rich CTT agar (top line). Growth on prey: ln-transformed population size estimates of M. xanthus (solid lines) and nine prey in the absence (dashed lines) and presence (dotted lines) of predator through time. The shaded area in each panel corresponds to the area bounded by M. xanthus population sizes on CTT and starvation agar controls. Error bars indicate the standard error of the mean.
Results
Trait variation across prey environments
Predator swarming
Myxococcus swarming rate on prey lawns served as our proxy for overall predatory performance and reflects the rate of population increase both while cells are dividing and while the colony is expanding via gliding motility. Prey-specific swarming rate varied greatly across prey species (Fig. 1, Table 2). Consistent with previous results [25], swarming was fastest on Escherichia coli, which supported faster swarm expansion than did CTT agar plates containing 1% Casitone as the growth substrate (Fig. 1, Wilcoxon rank sum test, p = 0.029). At the opposite extreme, M. xanthus failed to swarm more on C. citreum lawns than on prey-free TPM starvation agar (Fig. 1). This result is consistent with an earlier finding that C. citreum appears to inhibit swarming by most M. xanthus natural isolates relative to starvation agar controls [25]. Rhizobium vitris (RV) and Xanthomonas fragariae (XF) supported only low degrees of prey-induced swarming, whereas Bacillus bataviensis (BB), Arthrobacter globiformis (AG), Pseudomonas fluorescens (PF), Cytophaga johnsonae (CJ) and Micrococcus luteus (ML) supported prey-specific swarming rates intermediate between those on X. fragariae and E. coli (EC) (in order slowest to fastest, Fig. 1).
TABLE 2.
Variation within parameters and correlations between paired parameters.
| SR | GR | MCY | PKR | MPK | |
|---|---|---|---|---|---|
| SR | AOV p < 0.001 | rho = 0.383 | rho = 0.517 | rho = 0.685 | rho = 0.541 |
| KW p = 0.003 | p = 0.025 | p = 0.002 | p < 0.001 | p = 0.002 | |
| GR | r = 0.405 | AOV p < 0.001 | rho = 0.340 | rho = 0.268 | rho = 0.433 |
| p = 0.017 | KW p = 0.005 | p = 0.040 | p = 0.114 | p = 0.016 | |
| MCY | r = 0.463 | r = 0.345 | AOV p = 0.303 | rho = 0.573 | rho = 0.780 |
| p = 0.006 | p = 0.039 | KW p = 0.061 | p < 0.001 | p < 0.001 | |
| PKR | r = 0.621 | r = 0.258 | r = 0.565 | AOV p = 0.054 | rho = 0.507 |
| p < 0.001 | p = 0.128 | p < 0.001 | KW p = 0.049 | p = 0.004 | |
| MPK | r = 0.545 | r = 0.401 | r = 0.823 | r = 0.481 | AOV p = 0.017 |
| p = 0.002 | p = 0.025 | p < 0.001 | p = 0.006 | KW p = 0.079 |
SR: swarming rate; GR: growth rate of the predator; PKR: prey kill rate; MCY: log10 (maximum cell yield of the predator); MPK: maximum prey kill. Shaded cells show the significance levels of tests for parameter variation using two measures: Analysis of Variance (AOV) and Kruskal-Wallis Analysis of Variance (KW). The bottom left portion of the table shows Pearson product moment correlation statistics. The upper right corner of the table shows the Spearman’s rank correlation statistics.
Predator growth in the absence of swarm expansion
We also measured the rate of predator population growth on prey lawns when the perimeter of Myxococcus territory was held constant. This was accomplished by homogeneously spreading the mixture of the initial predator and prey populations across the entire surface of a prey lawn (or agar surface in the case of CTT and TPM controls), thus allowing no opportunity for subsequent horizontal swarm expansion beyond the area initially populated by predator cells. M. xanthus populations grew significantly on all prey species under these conditions (Fig. 2, Table 3), but growth rate varied substantially across prey types (ANOVA, p < 0.001; Fig. 2, 3a, Table 2).
TABLE 3.
Mean values and prey rank position for each parameter measured.
| Prey | SR | GR | MCY | PKR | MPK |
|---|---|---|---|---|---|
| AG | ***2.344 (5) | *0.093 (8) | ***3.95×1006 (2) | *0.019 (4) | * 0.122 (7) |
| BB | *2.323 (6) | **0.216 (2) | ***1.64×1006(6) | ns 0.011 (6) | *0.137 (5) |
| CC | ns −0.050 (9) | **0.039 (9) | ***4.35×1005 (9) | ns 0.001 (9) | ns 0.028 (9) |
| CJ | **2.729 (3) | *0.100 (6) | **1.52×1006 (7) | *0.009 (8) | • 0.069 (8) |
| EC | ***4.438 (1) | ***0.183 (3) | ***1.90×1006(5) | **0.034 (1) | ** 0.259 (1) |
| ML | **3.242 (2) | **0.165 (4) | **2.52×1006 (4) | *0.023 (3) | * 0.197 (3) |
| PF | **2.703 (4) | **0.223 (1) | ***4.23×1006(1) | *0.024 (2) | * 0.183 (4) |
| RV | **1.094 (8) | ***0.152 (5) | **9.47×1005 (8) | ns 0.013 (5) | • 0.215 (2) |
| XF | *1.547 (7) | **0.095 (7) | *2.75×1006(3) | ns 0.009 (7) | ** 0.133 (6) |
SR: Swarming rate; GR: Growth rate of the predator; MCY; Maximum cell yield attained by the predator; PKR: Prey kill rate; MPK: Maximum prey kill. Asterisks indicate significance levels for deviation from zero (SR, GR, PKR and MPK) or from the average yield of M. xanthus on nutrient-free TPM (MCY).
p < 0.001,
0.001 < p < 0.01,
0.01 < p < 0.05,
0.05 < p < 0.1;
ns p > 0.1.
The colors and numbers in parentheses both indicate ranks within a parameter (1 and green represent the highest rank, 9 and red represent lowest rank)
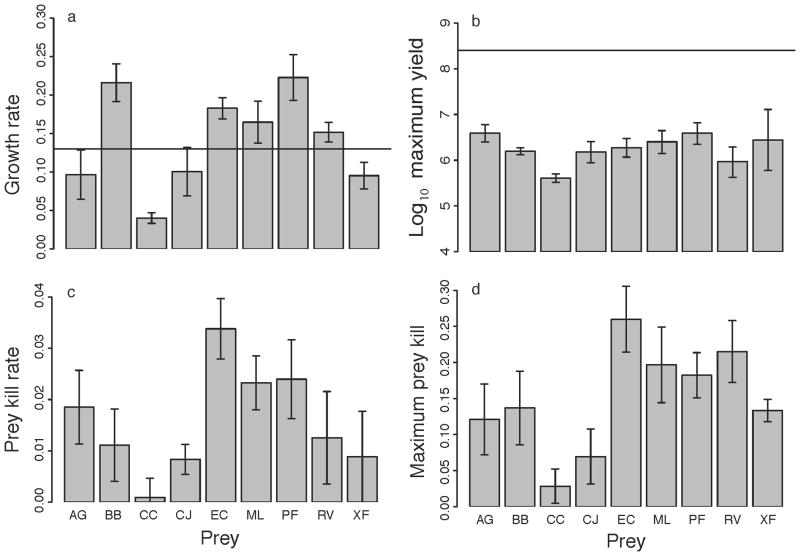
Figure 3.
Predator growth rate (a), maximum predator yield (b), prey kill rate (c) and maximum prey kill (d) for M. xanthus growth on nine prey species in the absence of swarm expansion. The solid lines in panels a and b represent the average value of each respective parameter measured on nutrient-rich CTT agar. The dashed line in panel b represents the average maximum yield on starvation agar. Error bars represent the standard error of the mean.
The inability of C. citreum to stimulate greater swarming by GJV2 than occurs on starvation agar control plates suggested that M. xanthus might be unable to convert C. citreum cells (or products) into utilizable growth substrate. However, in contrast to this expectation, M. xanthus was in fact able to grow significantly on C. citreum in the absence of swarm expansion well beyond the minimal growth that occurs on prey-free starvation agar plates (Fig. 2, 3a, Table 3).
Growth rate in the absence of swarm expansion correlated significantly with prey-specific swarming rate across prey types (Pearson correlation r = 0.41, p = 0.017; Table 2). Nonetheless, despite this overall correlation relative performance ranks during swarming vs. growth without swarming are reversed in several instances (Table 3). For example, M. xanthus swarms faster across lawns of A. globiformis, C. johnsonae and X. fragariae than across R. vitris but nonetheless grows faster on R. vitris in the absence of swarm expansion than it does on those three species (Figs. 1–3). Also, although E. coli supported the fastest swarming by M. xanthus on any prey (Fig. 1), growth rate on E. coli in the absence of swarm expansion is similar to or even lower than growth rate on several other prey (BB, ML, PF and RV; Fig. 3a).
The maximum cell yield attained by M. xanthus during growth on prey in the absence of swarm expansion was greater than that on starvation agar and less than that on CTT agar for all prey types (Fig. 3b) but did not vary significantly across prey types (ANOVA, p = 0.303; Table 2). Despite the non-significance of variation, this parameter nonetheless correlated significantly with prey-specific swarming rate, our primary measure of predatory performance (r = 0.46, p = 0.006; Table 2), as well as with growth rate in the absence of swarm expansion (r = 0.35, p = 0.039; Table 2). Consistent with our other assays indicating that predatory performance is weakest on C. citreum, this species supported the lowest maximum predator yield (Fig. 3b, Table 3).
Predator-induced prey death
The rate at which Myxococcus killed prey cells varied across prey species (ANOVA, p = 0.054; Fig. 3c, Table 2), as did the maximum number of prey cells killed by M. xanthus (ANOVA, p = 0.017; Fig. 3d, Table 2). Prey kill rate correlated significantly with overall prey-specific swarming rate (r = 0.62, p < 0.001; Table 2) but not with growth rate in the absence of swarm expansion (r = 0.26, p = 0.128; Table 2). Maximum kill correlated strongly with both prey-specific swarming rate (r = 0.55, p = 0.002; Table 2) and growth rate in the absence of swarm expansion (r = 0.40, p = 0.025; Table 2). The prey with highest (E. coli) and lowest (C. citreum) maximum kill values supported the fastest and slowest rates of M. xanthus swarming, respectively (Figs. 1, 3; Table 3).
Significant killing of prey by M. xanthus was detected for all prey species except C. citreum (Figs. 2, 3c, d; Table 3), which nonetheless supported M. xanthus growth (Figs. 2, 3a; Table 3). A likely reason why predator killing of C. citreum was not detected is that M. xanthus growth on C. citreum was slower than on any other prey (Table 3). The maximum population size achieved by M. xanthus on C. citreum by the end of our growth assays may have been too low to kill a detectable number of C. citreum cells (Fig. 2). In support of this hypothesis, significant predator-specific prey death for all other prey species was only detected after ≥60 hours, when M. xanthus had reached population sizes equal to or greater than the maximum size achieved on C. citreum at the final point of our growth assays. Detection of prey death lagged behind detection of M. xanthus growth for all prey, presumably because a minimum predator density had to be reached before prey death became detectable with our assay.
Intriguingly, M. xanthus colonies expand horizontally on C. citreum lawns no faster than on starvation agar (Fig. 1, Table 3, [25]). This poor performance at swarming on C. citreum is not attributable merely to a slow rate of cell division because on plates that did not allow swarm expansion, M. xanthus populations exhibited greater growth on C. citreum than on starvation agar (see later time points of CC graph in Fig. 2). Thus, C. citreum lawns appear to actually hinder the expansion of swarming M. xanthus colonies, possibly due to the secretion of an extracellular matrix that is physically difficult for M. xanthus cells to migrate through or that biochemically hinders motility functions.
Gram− vs. Gram+ differences
A previous study examining the predatory performance of many M. xanthus natural isolates across a large number of prey species found that, on average, M. xanthus isolates swarmed significantly faster on Gram– prey species than on Gram+ species [25]. We thus tested for any phylogenetically based differences in the average parameter values for these two prey categories (four Gram- and five Gram+ species). Gram– and Gram+ species were found to differ significantly only for maximum prey kill (Gram– mean = 0.106, Gram+ mean = 0.201, Wilcoxon rank sum test, p = 0.008).
Discussion
Much research has sought to characterize how the physical and behavioral traits of individuals contribute to animal predation [41–43]. For example, the two shrew species Neomys fodiens and Neomys anomalus co-exist in streams, but N. fodiens individuals are able to dive longer than N. anomalus and thus are able to reach and utilize a wider variety of prey [44, 45]. In contrast, fewer studies have sought to explore the relationships between the overall predatory success of a population and distinct population-level parameters that might be predictive of such success [19, 46–48].
Here we quantified several predatory parameters during growth of a motile bacterial predator on prey (growth rate in the absence of swarm expansion, maximum predatory yield, predator-induced prey death rate and maximum prey kill) and examined them both for variation across prey species and for their degree of correlation with a measure of overall predatory performance, namely prey-specific swarming rate across a prey lawn. Variation in each of these four alternative parameters across prey was found to be significantly predictive of prey-specific swarming rate on prey lawns.
These observed correlations among our population parameters may have implications for how these parameters evolve, whether in response to selective pressure imposed on M. xanthus by prey or due to other forces. Specifically, our results suggest causal relationships among the parameters and that substantial evolutionary changes in one will tend to be associated with corresponding changes in others. Hypothetically, for instance, predator ineffectiveness at killing prey cells might cause not only a slow rate of prey death, but also slow predator population growth and might be mechanistically related to how much growth substrate predator cells can extract prey cells and thus also affect maximum predatory yield. Nonetheless, despite the overall parameter correlations there were several instances in which the relative ranks of prey species varied greatly across parameters (Table 3), thus indicating that these parameters can evolve independently to some degree.
One striking example of rank difference across parameters is the comparison of M. xanthus swarming vs. non-swarming growth on E. coli. Although predator growth on E. coli is slightly slower than on two other prey in the absence of swarm expansion (Fig. 3a, P. fluorescens and B. bataviensis), predator swarming on E. coli – which involves both numerical increase by cell divisions and territorial expansion – is much faster than on all other prey (Fig. 1). Thus, E. coli somehow uniquely promotes territorial expansion in a manner independent of exponential predator growth rate within a non-expanding territory. In principle, this effect might be due to individual M. xanthus cells at the leading edge of a swarm dividing at a faster rate when they are free to migrate into an open field of E. coli cells than in the presence of other prey. Alternatively, E. coli cells might biologically stimulate and/or physically allow faster predator motility than do other prey and thus promote enhanced territorial expansion specifically by effects on motility. Another exception to the general correlations found here is C. johnsonae, which supports one of the fastest prey-specific swarming rates but ranks low for all four of the other parameters (Figs. 1, 3, Table 3). Also, M. xanthus swarm expansion is slower on R. vitris lawns than any other prey species except C. citreum but M. xanthus nonetheless appears to have killed a larger fraction of R. vitris populations than those of any prey other than E. coli (Figs. 1, 3d, Table 3).
The extent of evolutionary co-variance among the predation parameters examined here could be tested either with laboratory evolution experiments or comparative studies of divergent natural isolates. For example, M. xanthus lineages selected for improved predatory performance in a laboratory prey environment evolved to swarm more efficiently across agar prey arenas [19]. Such evolved lineages could be examined for whether predation parameters underwent changes in a manner consistent with the correlations found here and analysis of accumulated mutations in such lineages could provide mechanistic insights not revealed by our correlation approach.
Two of the parameters examined here (prey kill rate and maximum prey kill) reflect predator effects on prey populations. Thus, the extent of co-variance among these and other parameters may have community-level implications. For example, evolutionary changes in predator cell division rate or maximum yield may be causally linked to prey kill rate and consequently affect how prey populations change over time. This possibility highlights the importance of investigating the nature of predator-prey interactions for understanding the composition and dynamics of microbial communities.
Acknowledgments
This study was funded by a Doctoral Fellowship from Fundacao para a Ciencia e Tecnologia (SFRH / BD / 36183 / 2007) to H.M.-S. and NIH grant GM07690 to G.J.V.
References
- 1.Bruno JF, Cardinale BJ. Cascading effects of predator richness. Front Ecol Environ. 2008;6:539–546. [Google Scholar]
- 2.Drossel B, Higgs PG, McKane AJ. The influence of predator–prey population dynamics on the long-term evolution of food web structure. J Theor Biol. 2001;208:91–107. doi: 10.1006/jtbi.2000.2203. [DOI] [PubMed] [Google Scholar]
- 3.Hall SR, Duffy MA, Caceres CE. Selective predation and productivity jointly drive complex behavior in host-parasite systems. Am Nat. 2005;165:70–81. doi: 10.1086/426601. [DOI] [PubMed] [Google Scholar]
- 4.Brose U, Ehnes RB, Rall BC, Vucic-Pestic O, Berlow EL, Scheu S. Foraging theory predicts predator-prey energy fluxes. J Anim Ecol. 2008;77:1072–1078. doi: 10.1111/j.1365-2656.2008.01408.x. [DOI] [PubMed] [Google Scholar]
- 5.Petchey OL, Beckerman AP, Riede JO, Warren PH. Size, foraging, and food web structure. Proc Natl Acad Sci USA. 2008;105:4191–4196. doi: 10.1073/pnas.0710672105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmitz OJ. Effects of predator hunting mode on grassland ecosystem function. Science. 2008;319:952–954. doi: 10.1126/science.1152355. [DOI] [PubMed] [Google Scholar]
- 7.Corno G, Jurgens K. Direct and indirect effects of protist predation on population size structure of a bacterial strain with high phenotypic plasticity. Appl Environ Microbiol. 2006;72:78–86. doi: 10.1128/AEM.72.1.78-86.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guerrero R, Esteve I, Pedrós-Alió C, Gaju N. Predatory bacteria in prokaryotic communities - The earliest trophic relationships. Ann NY Acad Sci. 2006:1–13. [Google Scholar]
- 9.Velicer GJ, Mendes-Soares H. Bacterial predators. Curr Biol. 2009;19:R55–R56. doi: 10.1016/j.cub.2008.10.043. [DOI] [PubMed] [Google Scholar]
- 10.Gause GF, Smaragdova NP, Witt AA. Further studies of interaction between predators and prey. J Anim Ecol. 1936;5:1–18. [Google Scholar]
- 11.Luckinbill LS. Coexistence in laboratory populations of Paramecium aurelia and its predator Didinium nasutum. Ecology. 1973;54:1320–1327. [Google Scholar]
- 12.Jurkevitch E, editor. Predatory prokaryotes - Biology, Ecology and Evolution. Springer-Verlag; Heidelberg: 2007. [Google Scholar]
- 13.Casida LEJ. Minireview: Nonobligate bacterial predation of bacteria in soil. Microb Ecol. 1988;15:1–8. doi: 10.1007/BF02012948. [DOI] [PubMed] [Google Scholar]
- 14.Gaju N, Esteve I, Guerrero R. Distribution of predatory bacteria that attack Chromatiaceae in a sulfurous lake. Microb Ecol. 1992;24:171–179. doi: 10.1007/BF00174453. [DOI] [PubMed] [Google Scholar]
- 15.Rashidan KK, Bird DF. Role of predatory bacteria in the termination of cyanobacterial bloom. Microb Ecol. 2001;41:97–105. doi: 10.1007/s002480000074. [DOI] [PubMed] [Google Scholar]
- 16.McBride MJ, Zusman DR. Behavioral analysis of single cells of Myxococcus xanthus in response to prey cells of Escherichia coli. FEMS Microbiol Lett. 1996;137:227–231. doi: 10.1111/j.1574-6968.1996.tb08110.x. [DOI] [PubMed] [Google Scholar]
- 17.Rosenberg E, Keller KH, Dworkin M. Cell density-dependent growth of Myxococcus xanthus on casein. J Bacteriol. 1977;129:770–777. doi: 10.1128/jb.129.2.770-777.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holling CS. Principles of insect predation. Annu Rev Entomol. 1961;6:163–182. [Google Scholar]
- 19.Hillesland KL, Lenski RE, Velicer GJ. Ecological variables affecting predatory success in Myxococcus xanthus. Microb Ecol. 2007;53:571–578. doi: 10.1007/s00248-006-9111-3. [DOI] [PubMed] [Google Scholar]
- 20.Spormann AM. Gliding motility in bacteria: insights from studies of Myxococcus xanthus. Microbiol Mol Biol Rev. 1999;63:621–641. doi: 10.1128/mmbr.63.3.621-641.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hodgkin J, Kaiser D. Genetics of gliding motility in Myxococcus xanthus (Myxobacterales): two gene systems control movement. MGG. 1979;171:177–191. [Google Scholar]
- 22.Sudo S, Dworkin M. Bacteriolytic enzymes produced by Myxococcus xanthus. J Bacteriol. 1972;110:236–245. doi: 10.1128/jb.110.1.236-245.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weissman KJ, Müller R. A brief tour of myxobacterial secondary metabolism. Bioorg Med Chem. 2009;17:2121–2136. doi: 10.1016/j.bmc.2008.11.025. [DOI] [PubMed] [Google Scholar]
- 24.Berleman JE, Kirby JR. Deciphering the hunting strategy of a bacterial wolfpack. FEMS Microbiol Rev. 2009;33:942–957. doi: 10.1111/j.1574-6976.2009.00185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morgan AD, MacLean RC, Hillesland KL, Velicer GJ. Comparative analysis of Myxococcus predation on soil bacteria. Appl Environ Microbiol. 2010;76:6920–6927. doi: 10.1128/AEM.00414-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berleman JE, Chumley T, Cheung P, Kirby JR. Rippling is a predatory behavior in Myxococcus xanthus. J Bacteriol. 2006;188:5888–5895. doi: 10.1128/JB.00559-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nan B, Zusman DR. Uncovering the mystery of gliding motility in the Myxobacteria. Annu Rev Genet. 2011;45:21–39. doi: 10.1146/annurev-genet-110410-132547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berleman JE, Scott J, Chumley T, Kirby JR. Predataxis behavior in Myxococcus xanthus. Proc Natl Acad Sci USA. 2008;105:17127–17132. doi: 10.1073/pnas.0804387105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hillesland KL, Velicer GJ. Resource level affects relative performance of the two motility systems of Myxococcus xanthus. Microb Ecol. 2005;49:558–566. doi: 10.1007/s00248-004-0069-8. [DOI] [PubMed] [Google Scholar]
- 30.Pham VD, Shebelut CW, Diodati ME, Bull CT, Singer M. Mutations affecting predation ability of the soil bacterium Myxococcus xanthus. Microbiology. 2005;151:1865–1874. doi: 10.1099/mic.0.27824-0. [DOI] [PubMed] [Google Scholar]
- 31.Holling CS. Some characteristics of simple types of predation and parasitism. The Can Entomol. 1959;91:385–398. [Google Scholar]
- 32.Allee WC. Animal aggregations. The Quarterly Review of Biology. 1927;2:367–398. [Google Scholar]
- 33.Kadam SV, Velicer GJ. Variable patterns of density-dependent survival in social bacteria. Behav Ecol. 2006;17:833–838. [Google Scholar]
- 34.Klappenbach JA, Dunbar JM, Schmidt TM. rRNA Operon Copy Number Reflects Ecological Strategies of Bacteria. Appl Environ Microbiol. 2000;66:1328–1333. doi: 10.1128/aem.66.4.1328-1333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lenski RE, Rose MR, Simpson SC, Tadler SC. Long-term experimental evolution in Escherichia coli. I . Adaptation and divergence during 2000 generations. Am Nat. 1991;138:1315–1341. [Google Scholar]
- 36.Velicer GJ, Raddatz G, Keller H, Deiss S, Lans C, Dinkelacker I, Schuster SC. Comprehensive mutation identification in an evolved bacterial cooperator and its cheating ancestor. Proc Natl Acad Sci USA. 2006;103:8107–8112. doi: 10.1073/pnas.0510740103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaiser D. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc Natl Acad Sci USA. 1979;76:5952–595956. doi: 10.1073/pnas.76.11.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Velicer GJ, Kroos L, Lenski RE. Loss of social behaviors by Myxococcus xanthus during evolution in an unstructured habitat. Proc Natl Acad Sci USA. 1998;95:12376–12380. doi: 10.1073/pnas.95.21.12376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reasoner DJ, Geldreich EE. A new medium for the enumeration and subculture of bacteria from potable water. Appl Environ Microbiol. 1985;49:1–7. doi: 10.1128/aem.49.1.1-7.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bretscher AP, Kaiser D. Nutrition of Myxococcus xanthus, a fruiting myxobacterium. J Bacteriol. 1978;133:762–762768. doi: 10.1128/jb.133.2.763-768.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mittelbach GG. Efficiency and body size: a study of optimal diet and habitat use by bluegills. Ecology. 1981;62:1370–1386. [Google Scholar]
- 42.Preisser EL, Orrock JL, Schmitz OJ. Predator hunting mode and habitat domain alter nonconsumptive effects in predator-prey interactions. Ecology. 2007;88:2744–2751. doi: 10.1890/07-0260.1. [DOI] [PubMed] [Google Scholar]
- 43.Svanbäck R, Eklov P. Morphology dependent foraging efficiency in perch: a trade-off for ecological specialization? Oikos. 2003;102:273–284. [Google Scholar]
- 44.Mendes-Soares H, Rychlik L. Differences in swimming and diving abilities between two sympatric species of water shrews: Neomys anomalus and Neomys fodiens (Soricidae) J Ethol. 2008;27:317–325. [Google Scholar]
- 45.Rychlik L. Overlap of temporal niches among four sympatric species of shrews. Acta Theriol. 2005;50:175–188. [Google Scholar]
- 46.Beddington JR, Hassel MP, Lawton JH. The components of arthropod predation II. The predator rate of increase. J Anim Ecol. 1976;45:165–185. [Google Scholar]
- 47.Hassell MP, Lawton JH, Beddington JR. The components of arthropod predation: I. The prey death-rate. J Anim Ecol. 1976;145:135–164. [Google Scholar]
- 48.Heineman RH, Bull JJ. Testing optimality with experimental evolution: lysis time in a bacteriophage. Evolution. 2007;61:1695–1709. doi: 10.1111/j.1558-5646.2007.00132.x. [DOI] [PMC free article] [PubMed] [Google Scholar]