Figure 5. Macro-scale chromatin features and nuclear architecture.
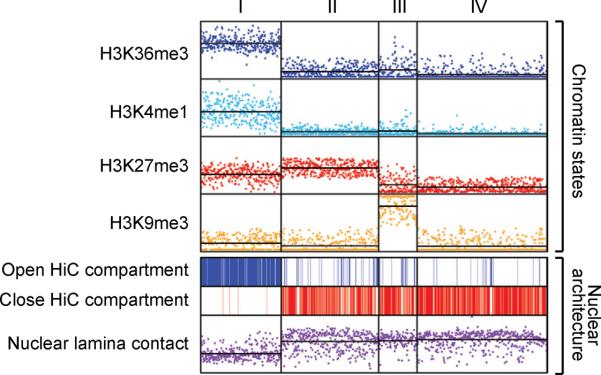
Non-overlapping 1 Mb genomic windows (n=2725) were clustered by their coverage by four histone modifications. Roughly half of the windows clustered into four main states: (I) `Active' loci with high H3K36me3 and H3K4me1; (II) `Polycomb-repressed' loci with high H3K27me3; (III) Heterochromatic loci with high H3K9me3; and (IV) `Null' loci devoid of histone modification. The top panel shows coverage for each interval (data points) and average coverage for each cluster (horizontal lines) by the indicated modification. The bottom panel indicates intervals that occupy active (top) or inactive (middle) nuclear compartments, and shows enrichment for nuclear lamina contacts (bottom). The data relate macro-scale modification patterns to genome compartmentalization and nuclear architecture.
