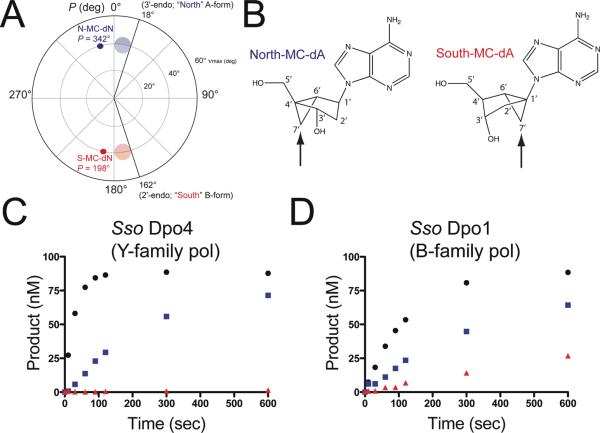
Figure 1.
Fixed-conformation nucleotide analogues probe the role of the furanose geometry during polymerase catalysis. (a) A pseudorotational cycle is shown to compare the N/S-MC conformations with the clustering of C3′-endo (semi-transparent blue circle) and C2′-endo sugar puckers (semi-transparent red circle) most commonly observed in RNA and DNA helices, respectively. (b) The chemical structures of N-MC-dA and S-MC-dA are shown with atoms in the ring system numbered. The arrows point to the different positioning of the cyclopropane ring relative to the cyclopentane ring. (c) Single-nucleotide insertion experiments were performed for Dpo4 (5 nM) and primer/template DNA (100 nM) with 10 μM dATP (black circles) N-MC-dATP (blue squares) and S-MC-dATP (red triangles). (d) Single-nucleotide insertion experiments were performed for Dpo1 (5 nM) and primer/template DNA (100 nM) with 10 μM dATP (black circles), N-MC-dATP (blue squares) and S-MC-dATP (red triangles).