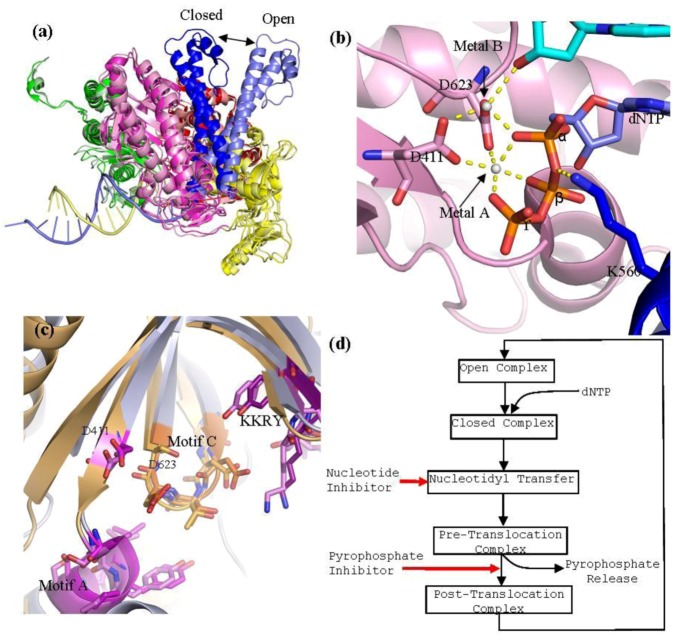
Figure 3.
(a) Superpositioning of open and closed structures of RB69 gp43 showing finger domain movement. This diagram is composed of RB69 gp43 in the fingers closed position (pdb 3LDS) [37] and in the fingers opened position (pdb 1IH7) [15]. (b) Polymerase active site of RB69 gp43 showing interactions between conserved residues of motif A and C, metal ions A and B and dNTP and interactions between K560 and dNTP. This image is an aligned composite image of pdb 3LDS [37] and 3SCX [38]. (c) Structural alignment of the polymerase active site of RB69gp43 (pdb 3LDS) and HSV1 UL30 (pdb 2GV9). RB69 gp43 backbone is in light blue while the HSV1 UL30 backbone is in light orange. Active site residues of RB69 gp43 are indicated. RB69 gp43 motif A is in magenta, motif C is in orange and KKRY is in purple. HSV1 UL30 motif A is in pink, motif C is in light orange and KKRY is in light purple. Images were generated using Pymol [23]. (d) Generalized diagram of the polymerase catalytic cycle showing steps at which inhibitors can act. Nucleotide inhibitors, once incorporated, prevent further extension of the DNA primer, by inhibiting nucleotidyl transfer. Whereas, pyrophosphate inhibitors mimicking the pyrophosphate leaving group, stabilizing the pre-translocation complex and prevent translocation.