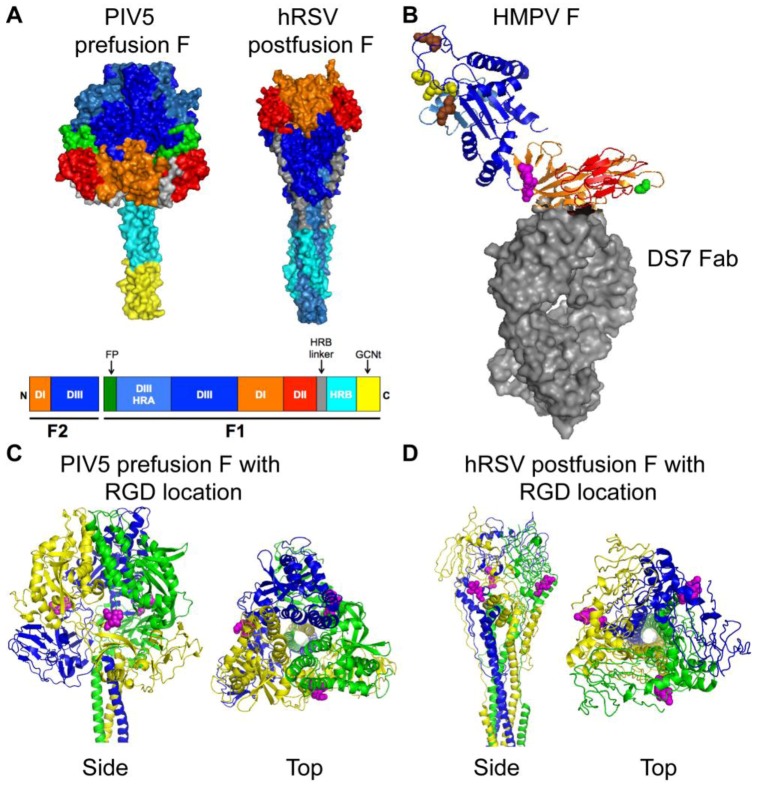
Figure 3.
Structures of paramyxovirus F proteins. (A) The prefusion conformation of the PIV5 F trimer and the postfusion conformation of the hRSV F trimer. A schematic of the F protein color-coded to identify structural domains is shown below the structures. The DI domain is shown in orange, the DII domain is shown in red, and the DIII domain is shown in blue. HRA is shown in light blue, HRB is shown in cyan and the HRB linker domain is shown in gray. The fusion peptide is shown in green for prefusion PIV5 F, but was not solved in the hRSV F structure. The GCNt domain (shown in yellow) was added to the PIV5 F trimer for crystallization. (B) HMPV F in complex with a neutralizing antibody (DS7 Fab, shown as gray surface). DI, DII, and DIII coloring is as for the F trimers shown in (A). The conserved RGD motif is shown as magenta spheres. Residue 294 (Glu) is shown as green spheres. Conserved charged residues in DIII that impact fusion activity are indicated as spheres (yellow, acidic residues; brown, basic residues). (C) Side and top views of the prefusion PIV5 F trimer are shown with the homologous residues to HMPV F-RGD shown as magenta spheres. Each monomer is shown in blue, yellow, or green. (D) Side and top views of the postfusion hRSV F trimer are shown with the homologous residues to HMPV F-RGD shown as magenta spheres. Coloring is as for PIV5 F in (C). Structural coordinates were obtained from the Protein Data Bank [52] and figure constructed with PyMOL. PIV5 F, PDB ID: 2B9B [53]; hRSV F, PDB ID: 3RKI [54]; HMPV F complex, PDB ID: 4DAG [55].