The homotrimeric protein YhbJ from E. coli has been homologously expressed, purified and crystallized in two distinct crystal forms. The crystals diffracted to 3.26 and 3.44 Å resolution.
Keywords: YhbJ, GlmYZ sRNA, GlmS, multi-angle laser light scattering, thermal shift assay, self-rotation function
Abstract
The protein YhbJ from Escherichia coli was previously reported to be involved in the regulation of glucosamine-6-phosphate synthase (GlmS) synthesis. YhbJ controls a regulatory cascade composed of the two small RNAs GlmY and GlmZ, which in turn regulate GlmS synthesis. For structural characterization, YhbJ was cloned, expressed and purified to homogeneity by Strep-tag affinity chromatography and size-exclusion chromatography. Multi-angle laser light-scattering analysis revealed its homotrimeric state in solution. The protein crystallized in two distinct trigonal crystal forms, with unit-cell parameters a = b = 91.62, c = 352.82 Å for space group P321 and a = b = 92.72, c = 156.75 Å for one of the enantiomorphic space groups P31 or P32. Preliminary analysis of the diffraction data suggests the presence of approximately three to seven molecules per asymmetric unit. Owing to the lack of a suitable homologous model, structure determination by means of MIR and MAD methods is required.
1. Introduction
Amino-sugars are essential building blocks in all living organisms. In bacteria, they are required for the synthesis of the cell-wall peptidoglycan and of lipopolysaccharides. The first step in amino-sugar synthesis is catalysed by the enzyme glucosamine-6-phosphate synthase (GlmS), which converts fructose-6-phosphate (e.g. from glycolysis) and glutamine to glucosamine-6-phosphate (GlcN-6-P) and glutamate (Milewski, 2002 ▶). GlcN-6-P thus represents the key intermediate in the biosynthesis of all amino-sugar macromolecules in the cell. However, when exogenous amino-sugars are available, these sugars are internalized and directly converted to GlcN-6-P, providing a bypass for the reaction catalysed by GlmS (Plumbridge & Vimr, 1999 ▶; Görke & Vogel, 2008 ▶). Since GlmS plays a crucial role in the biosynthetic pathway providing the building blocks for the cell-wall macromolecules, its activity must be tightly regulated. In Escherichia coli, synthesis of GlmS is regulated by a post-transcriptional negative-feedback mechanism that involves two homologous small RNAs (Kalamorz et al., 2007 ▶; Urban et al., 2007 ▶; Reichenbach et al., 2008 ▶; Urban & Vogel, 2008 ▶). These small RNAs (sRNAs), GlmY and GlmZ, share a sequence identity of 63% and provide the first example of a hierarchically acting regulatory cascade composed of two sRNAs (Reichenbach et al., 2008 ▶; Urban & Vogel, 2008 ▶). In addition, the protein YhbJ acts on these two RNAs in an as-yet unknown manner in order to modulate the expression of GlmS (Kalamorz et al., 2007 ▶).
Our current model for the feedback regulation of glmS expression by the sRNAs GlmY and GlmZ assumes that upon a decrease in the concentration of GlcN-6-P in the cell, the sRNA GlmY accumulates and acts via YhbJ to inhibit processing and degradation of the second sRNA GlmZ by its corresponding RNase (Kalamorz et al., 2007 ▶; Reichenbach et al., 2008 ▶; Urban & Vogel, 2008 ▶; Göpel & Görke, 2012 ▶). Thus, full-length GlmZ accumulates and, assisted by the RNA chaperone Hfq, can base-pair with the glmS mRNA transcript (Kalamorz et al., 2007 ▶; Urban & Vogel, 2008 ▶; Salim et al., 2012 ▶). The GlmZ–glmS interaction stimulates GlmS synthesis, which refills the GlcN-6-P pool (Kalamorz et al., 2007 ▶; Urban & Vogel, 2008 ▶). The homologous sRNA GlmY counteracts degradation of GlmZ presumably by sequestration of YhbJ. Thus, GlmY and GlmZ may compete for binding to YhbJ. Since this pathway is induced when the concentration of the enzymatic product GlcN-6-P of GlmS decreases, GlmY and GlmZ establish a feedback regulation that adjusts GlmS synthesis to the concentration of its product at a post-transcriptional level (Kalamorz et al., 2007 ▶; Urban & Vogel, 2008 ▶).
YhbJ does not display any notable similarity to other proteins of known function but it is highly conserved in Gram-positive and Gram-negative bacteria suggesting a crucial role in bacterial physiology (Pompeo et al., 2011 ▶). It is about 30 kDa in size and contains an N-terminal Walker A/B motif which allows ATP/GTP binding and hydrolysis (Luciano et al., 2009 ▶; Fig. 1 ▶ a). In addition, sequence analysis using the BindN server predicts putative RNA binding in YhbJ homologues of Gram-negative bacteria that also possess the GlmYZ sRNAs (Kalamorz et al., 2007 ▶; Wang & Brown, 2006 ▶). Accordingly, it is anticipated that YhbJ binds GlmYZ sRNAs. Since YhbJ does not display a definite RNA-binding motif with similarity to other structurally known motifs, the structure and mode of RNA binding are of great interest. Thus, YhbJ is probably the key regulator protein within the GlmYZ cascade which directs and controls the fate of the sRNAs, dependent on the GlcN-6-P status of the cell.
Figure 1.
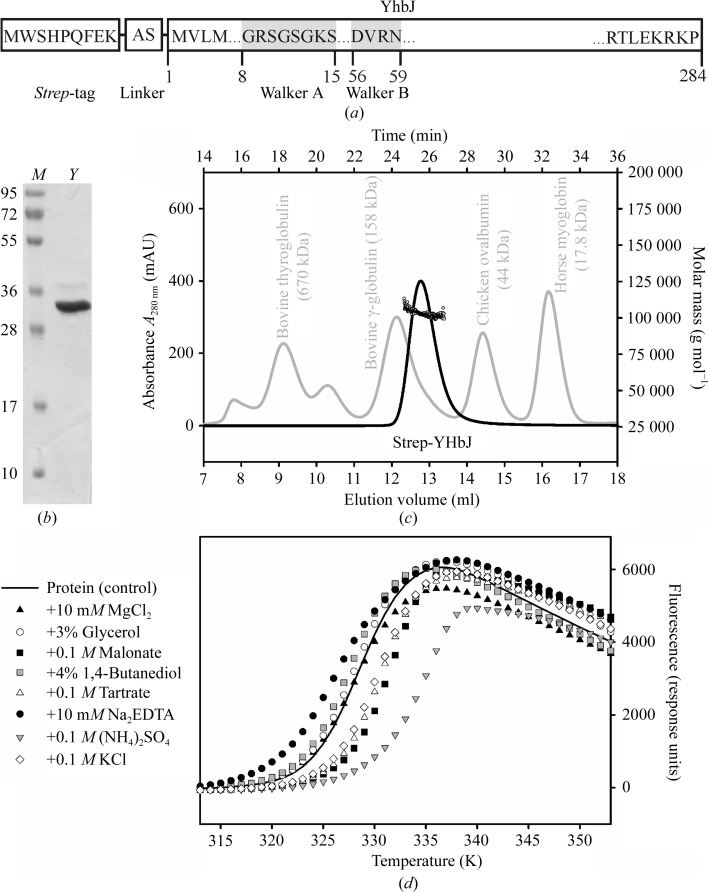
(a) The construct of YhbJ from E. coli used in this study. The protein contains a non-cleavable Strep-tag at its N-terminus followed by a short linker (Lüttmann et al., 2012 ▶). The YhbJ amino-acid sequence is indicated by the numbering 1–284. The ATP/GTP-binding motif is indicated by grey boxes (Luciano et al., 2009 ▶). (b) 15% SDS–PAGE of purified Strep-YhbJ stained with Coomassie Brilliant Blue. The molecular weight of the Strep-YhbJ monomer is 33.8 kDa (lane Y). Lane M contains molecular-weight markers (labelled in kDa). (c) SEC–MALLS: analytical size-exclusion chromatography of Strep-YhbJ (black line) together with reference proteins (grey line) on a Superdex 200 10/300 column coupled with multi-angle laser light scattering (black circles). The molar mass of Strep-YhbJ was determined to be 103 100 g mol−1 (± 0.9%), which corresponds to 3.1 times the molecular weight of the monomer. (d) Thermal shift assay for Strep-YhbJ in combination with the Hampton Additive Screen. For clarity, only selected conditions are presented. In comparison to Strep-YhbJ without any additives (control; solid line), ammonium sulfate led to a considerable shift in the apparent T m and thus could be identified as a highly stabilizing additive.
In this report, we describe the expression, purification and crystallization of full-length YhbJ from E. coli and the preliminary X-ray diffraction analysis of YhbJ crystals. Our data also indicate that the biological unit is a homotrimer in solution. This work represents the first step towards a full structural and functional characterization of YhbJ.
2. Experimental methods
2.1. Strains and plasmids
Plasmid pBGG164 (Lüttmann et al., 2012 ▶) was used for overproduction of Strep-YhbJ. This plasmid is a pBR322 derivative and carries the recombinant strep-yhbJ gene under control of the IPTG-inducible Ptac promoter, the lacIq gene for control of the Ptac promoter and the bla ampicillin-resistance gene (Lüttmann et al., 2012 ▶). Strep-tagged YhbJ was overproduced in E. coli strain Z106 that is deficient in the genes of glmY and glmZ. Z106 is a derivative of strain Z105 (Reichenbach et al., 2008 ▶), but has been cured from the cat resistance gene using helper plasmid pCP20 as previously described (Datsenko & Wanner, 2000 ▶). The construct of YhbJ from E. coli which we used is schematically depicted in Fig. 1 ▶(a).
2.2. Expression and purification
Cells were grown in Terrific broth medium in the presence of 100 µg ml−1 ampicillin at 310 K and 180 rev min−1. At an OD600nm of 0.8, the temperature was lowered to 289 K and protein expression was induced with 1 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG). After 16 h, the cells were harvested by centrifugation at 5000g and 277 K for 20 min and the pellet was stored at 193 K. For protein purification, 5 g of the pellet were resuspended in 20 mM HEPES buffer pH 7.8 containing 300 mM KCl, 2 mM MgCl2, 1 mM phenylmethanesulfonylfluoride (PMSF), 0.5 mg ml−1 lysozyme and 0.05 mg ml−1 RNase A. Cell rupture was carried out by passing the cells five times through a microfluidizer (Microfluidics). After 1 h centrifugation at 75 000g and 277 K, the supernatant was filtered through a 0.45 µm filter (Millipore) and subsequently loaded onto a 15 ml Strep-Tactin column (IBA Life Sciences; Schmidt & Skerra, 2007 ▶), pre-equilibrated with binding buffer consisting of 20 mM HEPES buffer pH 7.8, 300 mM KCl, 2 mM MgCl2. Strep-YhbJ was eluted with 2.5 mM desthiobiotin in binding buffer. Fractions containing the target protein were pooled and subsequently loaded onto a size-exclusion chromatography column (HiLoad Superdex 200 16/60, GE Healthcare), pre-equilibrated with 20 mM HEPES pH 7.5, 150 mM KCl, 2 mM MgCl2, in order to remove aggregates and protein impurities. The eluted protein was concentrated to 1.5–2.5 mg ml−1 using centrifugal filters (molecular-weight cutoff 30 000, Millipore). The protein was >95% pure as assessed by 15% SDS–PAGE with Coomassie Brilliant Blue staining (Laemmli, 1970 ▶) and was immediately used or stored at 277 K for no longer than 4 d. Protein concentrations were determined by the Bradford method using Bio-Rad protein assay dye reagent with bovine serum albumin as a standard (Bradford, 1976 ▶).
2.3. Multi-angle laser light scattering
0.2 ml Strep-YhbJ solution was loaded onto a 24 ml analytical size-exclusion column (Superdex 200 10/300 GL, GE Healthcare) at a concentration of 1 mg ml−1 and a flow rate of 0.5 ml min−1. The column was pre-equilibrated with 20 mM HEPES pH 7.5, 150 mM KCl, 2 mM MgCl2. Multi-angle laser light-scattering analysis was performed continuously on the column eluate at 291 K (size-exclusion chromatography coupled with multi-angle laser light scattering, SEC–MALLS). For detection, a miniDAWN TREOS triple-angle light-scattering detector (Wyatt Technology Corp.) was used. The data were processed with ASTRA 5.3.4.14 software (Wyatt Technology Corp.) employing a native extinction coefficient of ∊280nm = 22 900 M−1 cm−1 for the Strep-YhbJ protein. A theoretical molecular weight of 33.8 kDa for the monomer was later used as reference for calculation of the oligomeric state.
2.4. Thermal shift assay
Samples were diluted in 96-microplate wells (clear Multiplate 96-well PCR plates, Bio-Rad) to a final concentration of 1 µM Strep-YhbJ in 20 µl total volume. The samples consisted of 20 mM HEPES pH 7.5, 150 mM KCl, 2 mM MgCl2, 2× SYPRO Orange (diluted from a 5000× SYPRO Orange preparation from Molecular Probes) and 2 µl of Hampton Additive Screen (Hampton Research). The plates were then centrifuged at 5000g for 5 min at 277 K and sealed with optical-quality sealing tape (Bio-Rad). Subsequently, the samples were subjected to thermal denaturation in a Real Time PCR cycler equipped with a CFX96 optical reaction module (Bio-Rad) by applying a temperature gradient from 293 to 368 K and a ramping rate of 1 K min−1. Protein unfolding was monitored by the increase in the fluorescence of the SYPRO Orange probe, which was recorded every minute using excitation and emission wavelengths of 492 and 516 nm, respectively. The relative fluorescence emission intensity (response units) was plotted as a function of the temperature and the T m for each individual sample was estimated as the temperature corresponding to the midpoint between the baseline and the point with maximum fluorescence intensity (Ericsson et al., 2006 ▶).
2.5. Protein crystallization
Freshly purified Strep-YhbJ at concentrations between 1.5 and 2.5 mg ml−1 in 20 mM HEPES pH 7.5, 150 mM KCl, 2 mM MgCl2 was subjected to JBScreen Classic 1, 2, 4, 5, 6, 7, 8 and 10 (Jena Bioscience) as initial screening kits. Crystallization trials were performed using a Phoenix protein crystallization robot (Art Robbins Instruments) using the sitting-drop vapour-diffusion method on 96-well plates (SWISSCI MRC 3-well Crystallization Plate, Jena Bioscience). A mixture of 0.25 µl protein solution and 0.25 µl reservoir solution (or alternatively 0.125 µl reservoir solution) was equilibrated against 45 µl reservoir solution. Microcrystals were obtained in 1.0 M ammonium sulfate at 277 K. These conditions were optimized using the sitting-drop vapour-diffusion method in 24-well and 96-well plates. Crystallization parameters optimized included the concentration of the precipitant, the buffer substance, the pH and the use of various types of additives (Hampton Additive Screen). Crystals suitable for X-ray diffraction analysis were obtained by mixing 2 µl protein solution (1.5–2.5 mg ml−1 in 20 mM HEPES pH 7.5, 150 mM KCl, 2 mM MgCl2) with 1 µl reservoir solution [0.1 M MES, 0.8–1.1 M ammonium sulfate, 0.01 M β-mercaptoethanol, 1%(v/v) glycerol pH 6.7–7.0] and equilibrating against 0.7 ml reservoir solution at 277 K. The pH of the reservoir solution was adjusted after addition of ammonium sulfate. Crystals having a hexagonal shape grew to maximum dimensions of 0.15 × 0.15 × 0.05 mm after 4–10 d.
2.6. Diffraction data collection and processing
Before flash-cooling in liquid nitrogen, Strep-YhbJ crystals were soaked for 10 s in cryoprotectant solution that consisted of reservoir solution to which 20–25%(v/v) (2R,3R)-(−)2,3-butandiol had been added. Reservoir solutions supplemented with different concentrations of glycerol, various PEGs, sucrose and glucose were also tested for cryoprotection of Strep-YhbJ crystals but led to inferior diffraction behaviour. Native diffraction data sets from single Strep-YhbJ crystals were collected at 100 K on BL14.1 operated by the Helmholtz-Zentrum Berlin (HZB) at the BESSY II electron storage ring, Berlin-Adlershof, Germany (Mueller et al., 2012 ▶) and on microfocus beamline ID23-2 of the ESRF, Grenoble, France. Data for crystals belonging to space group P321 were collected at a wavelength of 0.91841 Å in 0.1° oscillation increments covering a total rotation range of 88°. The small oscillation angle per image was necessary to avoid partial overlaps of the spots. Data for crystals belonging to space group P31/P32 were collected at a wavelength of 0.87260 Å in 2.0° oscillation steps covering a total rotation range of 110°.
The strategy function in iMOSFLM (Battye et al., 2011 ▶) was used to reduce overlap as well as to maximize data completeness. The data sets were integrated and scaled using the programs XDS and XSCALE (Kabsch, 2010 ▶). Data were further processed using POINTLESS (Evans, 2006 ▶) and phenix.xtriage (Adams et al., 2010 ▶). Self-rotation functions were calculated using GLRF (Tong & Rossmann, 1997 ▶).
3. Results and discussion
Homologous expression of full-length YhbJ comprising a noncleavable Strep-tag at its N-terminus resulted in predominantly soluble protein. The recombinant protein expressed from the GlmYZ sRNA-deficient E. coli strain Z106 was purified in a two-step procedure applying Strep-Tactin affinity and size-exclusion chromatography to give a final yield of approximately 2 mg highly pure protein per litre of culture medium (Fig. 1 ▶ b). In order to investigate the oligomeric state of Strep-YhbJ, the purified sample was analysed by analytical SEC–MALLS. The sample showed a monomodal particle size distribution with a molar mass of 103 100 g mol−1 (± 0.9%; Fig. 1 ▶ c), corresponding to 3.1 times the molecular weight of the monomer (33.8 kDa). This finding suggests that the biological unit consists of a homotrimer in solution, which is also in accordance with the results from the analytical size-exclusion chromatography. Using bovine thyroglobulin (670 kDa), bovine γ-globulin (158 kDa), chicken ovalbumin (44 kDa) and horse myoglobin (17.8 kDa) as reference proteins, the apparent molecular weight of Strep-YhbJ was calculated to be 105.1 kDa, which also corresponds to 3.1 times the molecular weight of the monomer (Fig. 1 ▶ c).
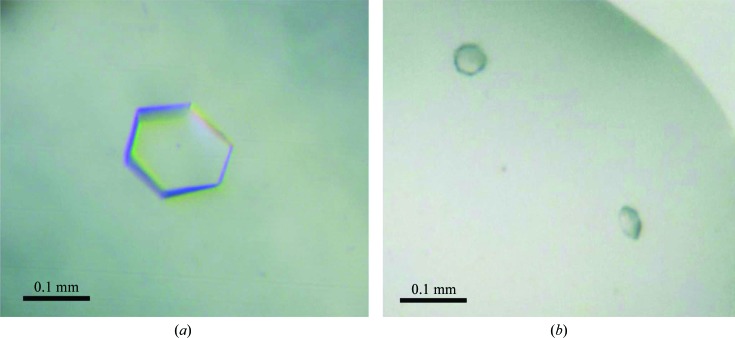
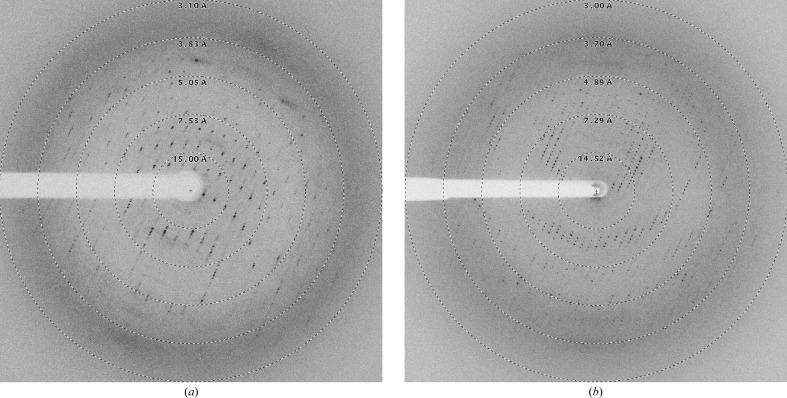
High-throughput crystallization screening using only freshly purified protein resulted in spherulite crystals in 1.0 M ammonium sulfate (condition A2 from JBScreen Classic 6). A favourable effect of ammonium sulfate on Strep-YhbJ was also confirmed by a thermal shift assay testing 96 different additives. This method has been described previously to be very helpful in optimizing protein crystallization and can serve as a powerful tool to predict crystallization conditions as in this case (Ericsson et al., 2006 ▶). Here, the apparent T m of the protein was shifted considerably to higher temperature, i.e. from 328.1 K without additive to 333.8 K in the presence of 0.1 M ammonium sulfate, indicating a significant stabilization of the protein (Fig. 1 ▶ d). Because sulfate is known to be a phosphate mimic, it is not surprising that YhbJ, which is anticipated to bind RNA and ATP/GTP, can be stabilized by sulfate. Moreover, high sulfate concentrations might ensure that the phosphate-binding sites of YhbJ are fully saturated, thus leading to the high degree of homogeneity needed for crystallization. A less pronounced stabilization effect is observed with the organic anions malonate and tartrate, which may result from interactions with positively charged amino-acid residues (Fig. 1 ▶ d). Upon further optimization of the initial crystallization condition, protein crystals suitable for X-ray data collection were obtained from ammonium sulfate with the additives glycerol and β-mercaptoethanol by mixing the protein and reservoir solutions in a 2:1 ratio (Fig. 2 ▶). The buffer pH was best at the theoretical pI of the protein (pI = 6.77). However, the crystals did not diffract X-ray radiation beyond 3.26 Å resolution (Fig. 3 ▶). Moreover, the protein crystallized in two distinct crystal forms which could not be easily distinguished under the microscope. Analysis of the pattern of systematic absences indicated that the crystals belong to the primitive trigonal space group P321 and to one of the enantiomorphic space groups P31/P32 (data statistics are shown in Table 1 ▶). Further validation of the space groups was performed by POINTLESS (Evans, 2006 ▶) and by manual inspection of the systematic absences, which confirmed the space groups with a systematic absence probability of 94.4% for P321 and 98.0% for P31/P32. In general, crystals of space group P321 grew to slightly larger dimensions than those of space group P31/P32. In addition, crystals of the latter space group turned out to be more sensitive to changes in mother-liquor composition. Small changes arising from cryosoaking or heavy-atom soaking easily resulted in substantial decay of the diffraction power.
Figure 2.
Two distinct crystal forms of Strep-YhbJ. (a) Strep-YhbJ crystal belonging to space group P321. (b) Strep-YhbJ crystals belonging to one of the enantiomorphic space groups P31/P32. Both hexagonal-shaped crystal forms crystallized under the same conditions.
Figure 3.
X-ray diffraction images of Strep-YhbJ crystals belonging to space groups P321 (a) and P31/P32 (b).
Table 1. Data-collection and processing statistics.
Values in parentheses are for the outer resolution shell.
| Beamline | BESSY BL14.1 | ESRF ID23-2 |
| Wavelength (Å) | 0.91841 | 0.87260 |
| Detector | MX-225 | MAR/Rayonix 3 × 3 Mosaic 225 |
| Crystal-to-detector distance (mm) | 365.57 | 374.34 |
| Space group | P321 | P31 or P32 |
| Rotation range per image (°) | 0.1 | 2.0 |
| Total rotation range (°) | 88 | 110 |
| No. of crystals | 1 | 1 |
| Unit-cell parameters (Å, °) | a = b = 91.62, c = 352.82, α = β = 90.0, γ = 120.0 | a = b = 92.72, c = 156.75, α = β = 90.0, γ = 120.0 |
| Solvent content (%) | 72–35† | 70–28† |
| Matthews coefficient V M (Å3 Da−1) | 4.4–1.9† | 4.0–1.7† |
| Molecules per asymmetric unit | 3–7† | 3–7† |
| Resolution range (Å) | 44.34–3.26 (3.43–3.26) | 46.36–3.44 (3.59–3.44) |
| Unique reflections | 27708 (3833) | 19961 (2382) |
| Average multiplicity | 4.67 (4.77) | 3.60 (3.58) |
| Completeness (%) | 99.1 (99.8) | 99.8 (99.9) |
| Mean I/σ(I) | 13.37 (2.56) | 9.87 (1.92) |
| R merge ‡ (%) | 8.6 (76.2) | 12.0 (63.8) |
| R meas § (%) | 9.7 (85.5) | 14.1 (75.2) |
| CC1/2 ¶ (%) | 99.8 (76.8) | 99.4 (69.5) |
| Estimated mosaicity (°) | 0.11 | 0.22 |
| Wilson B value (Å2) | 84.8 | 71.8 |
| Estimated twin fraction (%) | 6.2 | 4.9 |
| Twin law (merohedral twinning) | −h, −k, +l | +k, +h, −l |
The nature of the asymmetric unit has not yet been clarified, but it is expected to contain between three and seven monomers of Strep-YhbJ.
R
merge = 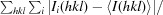
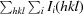 .
.
R
meas = 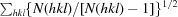
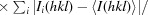
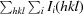 , where Ii(hkl) is the ith observation of reflection hkl, 〈I(hkl)〉 is the weighted average intensity for all observations i of reflection hkl and N is the number of observations of reflection hkl.
, where Ii(hkl) is the ith observation of reflection hkl, 〈I(hkl)〉 is the weighted average intensity for all observations i of reflection hkl and N is the number of observations of reflection hkl.
CC1/2 is the correlation coefficient between intensities from random halves of the data set as reported by XSCALE (Kabsch, 2010 ▶).
In order to determine the number of Strep-YhbJ molecules in the asymmetric unit, Matthews coefficients (V M) were calculated for both crystal forms (Matthews, 1968 ▶). Based on the total molecular weight of 33.8 kDa and the unit-cell parameters, V M calculation indicated that the asymmetric units of both crystal lattices theoretically contain between three and seven monomers. These values are equivalent to solvent contents between 72 and 28%, and lie within the normal range of values observed for soluble protein crystals (Matthews, 1968 ▶). However, the weak scattering behaviour of the crystals suggests that the solvent content is quite high and that the asymmetric unit therefore only contains about three to four molecules. To evaluate the point-group symmetry of the crystallized protein, self-rotation functions were calculated in reciprocal space using polar angles as implemented in GLRF (Tong & Rossmann, 1997 ▶). A strong peak appears parallel to the crystallographic c axis in the κ = 120° section at ψ = 90°, ϕ = 90°, which is characteristic for both space groups P321 and P31/P32 (Sawaya, 2007 ▶). In addition, for space group P321 (but not for P31/P32) we observe perpendicular to it, in the κ = 180° section, characteristic peaks at ψ = 30°/ϕ = 0°, ψ = 90°/ϕ = 0° and ψ = 150°/ϕ = 0°. All these peaks are consistent with their respective space group and thus the self-rotation function did not indicate any peaks corresponding to noncrystallographic symmetry (Sawaya, 2007 ▶). The largest off-origin peaks in the native Patterson map were 3.1% (P321) and 3.7% (P31/P32) of the height of the origin peak, which also suggested no significant pseudo-translation. If we want to make use of our observation that YhbJ is a homotrimer in solution and that also the crystals display threefold symmetry, it is tempting to postulate the existence of one or two trimers per asymmetric unit. Thus, four, five and seven molecules per asymmetric unit would be very unlikely, especially for space group P31/P32, unless we assume that the protein does not form a trimer in the crystal. However, since we cannot exclude the possibility that one or two trimer(s) are generated by crystallographic symmetry in P321, for instance when a trimer axis hides its existence behind the crystallographic symmetry axis in the self-rotation plot, the exact number of molecules can only be answered by the final structure determination.
The structure of Strep-YhbJ could not be determined by molecular replacement. Except for a low sequence identity to some structurally known kinases (12–21%; Jaroszewski et al., 2011 ▶), searches of the PDB using the YhbJ amino-acid sequence failed to identify any usable template. We are currently working on preparing crystals of heavy-atom derivatives and attempting to solve the structure using multiple isomorphous replacement methods. In addition, selenomethionine-substituted protein crystals were prepared for multiple-wavelength anomalous dispersion with crystals grown under the same conditions as for the native protein.
Acknowledgments
We would like to thank Michael Franke for setting up the crystallization screens. We are grateful for the support of the BESSY II (Berlin-Adlershof, Germany) and the ESRF (Grenoble, France) and would like to thank the beamline staff for assistance during data collection. Piotr Neumann is acknowledged for helpful advice and technical assistance.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Battye, T. G. G., Kontogiannis, L., Johnson, O., Powell, H. R. & Leslie, A. G. W. (2011). Acta Cryst. D67, 271–281. [DOI] [PMC free article] [PubMed]
- Bradford, M. M. (1976). Anal. Biochem. 72, 248–254. [DOI] [PubMed]
- Datsenko, K. A. & Wanner, B. L. (2000). Proc. Natl Acad. Sci. USA, 97, 6640–6645. [DOI] [PMC free article] [PubMed]
- Ericsson, U. B., Hallberg, B. M., DeTitta, G. T., Dekker, N. & Nordlund, P. (2006). Anal. Biochem. 357, 289–298. [DOI] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Göpel, Y. & Görke, B. (2012). Curr. Opin. Microbiol. 15, 132–139. [DOI] [PubMed]
- Görke, B. & Vogel, J. (2008). Genes Dev. 22, 2914–2925. [DOI] [PubMed]
- Jaroszewski, L., Li, Z., Cai, X., Weber, C. & Godzik, A. (2011). Nucleic Acids Res. 39, W38–W44. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kalamorz, F., Reichenbach, B., März, W., Rak, B. & Görke, B. (2007). Mol. Microbiol. 65, 1518–1533. [DOI] [PubMed]
- Laemmli, U. K. (1970). Nature (London), 227, 680–685. [DOI] [PubMed]
- Luciano, J., Foulquier, E., Fantino, J. R., Galinier, A. & Pompeo, F. (2009). J. Bacteriol. 191, 1556–1564. [DOI] [PMC free article] [PubMed]
- Lüttmann, D., Göpel, Y. & Görke, B. (2012). Mol. Microbiol. 86, 96–110. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Milewski, S. (2002). Biochim. Biophys. Acta, 1597, 173–192. [DOI] [PubMed]
- Mueller, U., Darowski, N., Fuchs, M. R., Förster, R., Hellmig, M., Paithankar, K. S., Pühringer, S., Steffien, M., Zocher, G. & Weiss, M. S. (2012). J. Synchrotron Rad. 19, 442–449. [DOI] [PMC free article] [PubMed]
- Plumbridge, J. & Vimr, E. (1999). J. Bacteriol. 181, 47–54. [DOI] [PMC free article] [PubMed]
- Pompeo, F., Luciano, J., Brochier-Armanet, C. & Galinier, A. (2011). J. Mol. Microbiol. Biotechnol. 20, 156–167. [DOI] [PubMed]
- Reichenbach, B., Maes, A., Kalamorz, F., Hajnsdorf, E. & Görke, B. (2008). Nucleic Acids Res. 36, 2570–2580. [DOI] [PMC free article] [PubMed]
- Salim, N. N., Faner, M. A., Philip, J. A. & Feig, A. L. (2012). Nucleic Acids Res. 40, 8021–8032. [DOI] [PMC free article] [PubMed]
- Sawaya, M. R. (2007). Methods Mol. Biol. 364, 95–120. [DOI] [PubMed]
- Schmidt, T. G. & Skerra, A. (2007). Nature Protoc. 2, 1528–1535. [DOI] [PubMed]
- Tong, L. & Rossmann, M. G. (1997). Methods Enzymol. 276, 594–611. [PubMed]
- Urban, J. H., Papenfort, K., Thomsen, J., Schmitz, R. A. & Vogel, J. (2007). J. Mol. Biol. 373, 521–528. [DOI] [PubMed]
- Urban, J. H. & Vogel, J. (2008). PLoS Biol. 6, e64. [DOI] [PMC free article] [PubMed]
- Wang, L. & Brown, S. J. (2006). Nucleic Acids Res. 34, W243–W248. [DOI] [PMC free article] [PubMed]