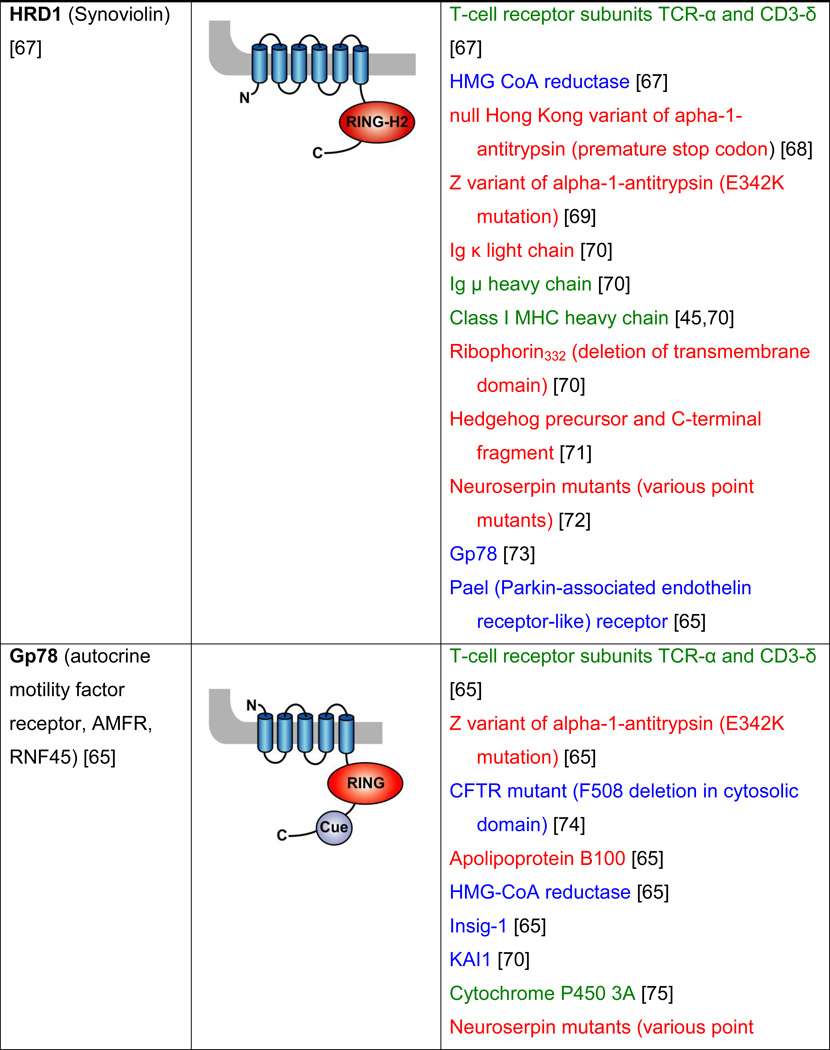
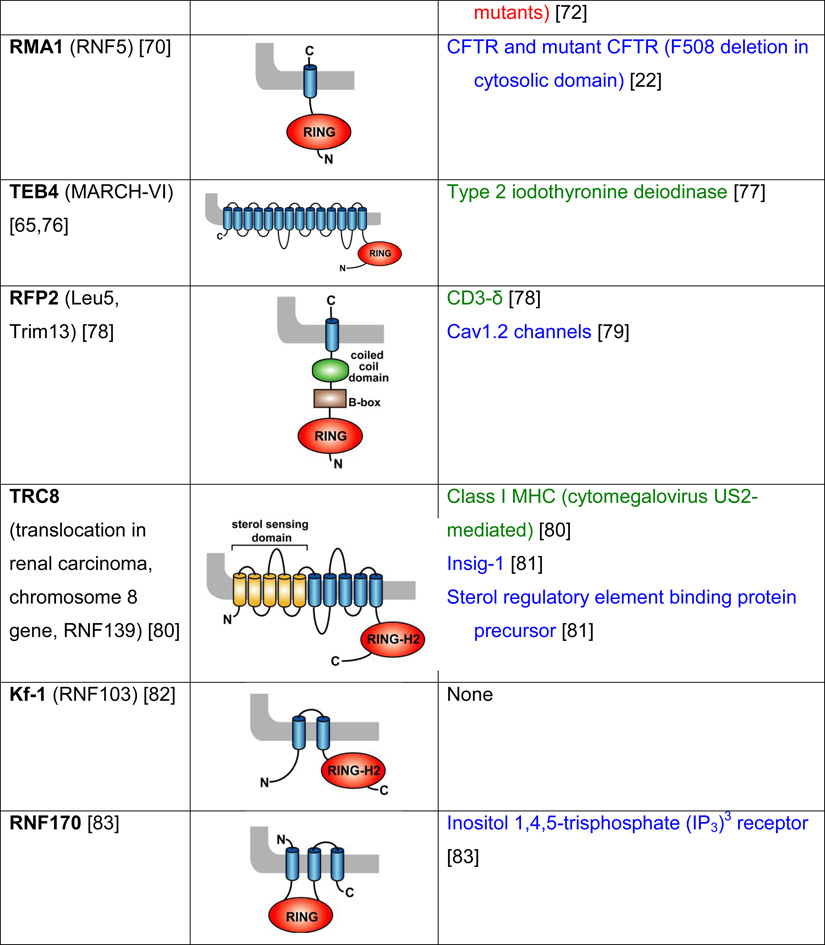
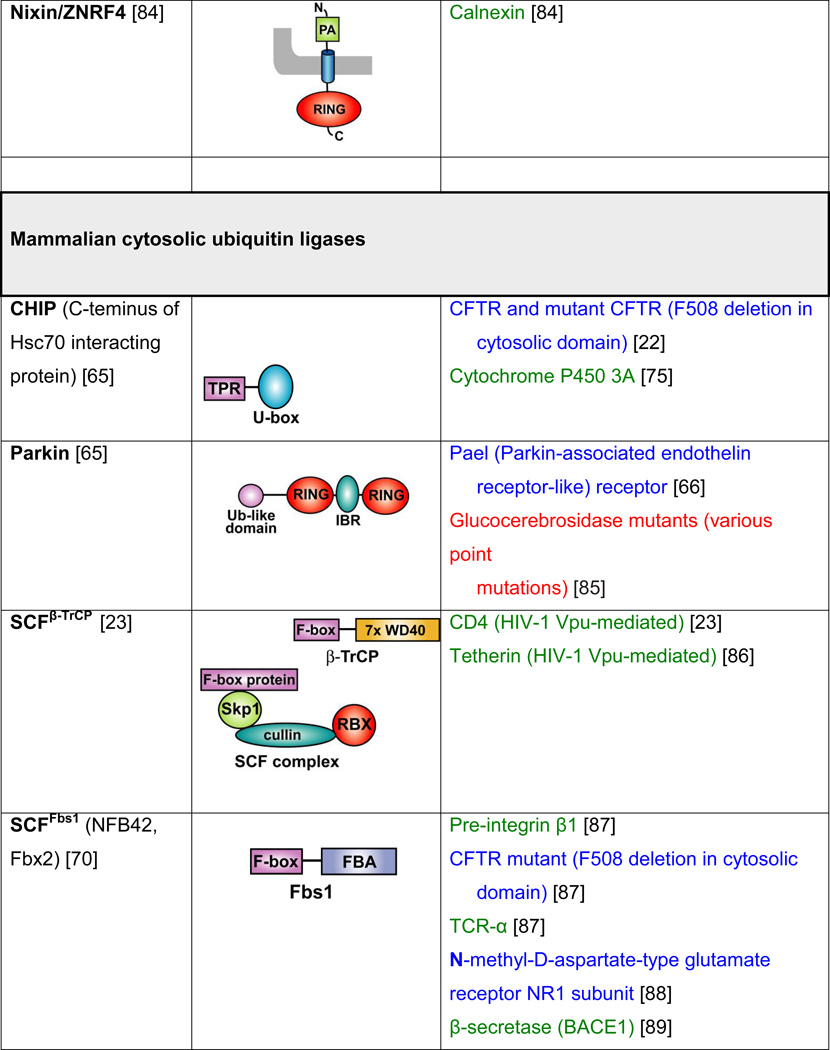
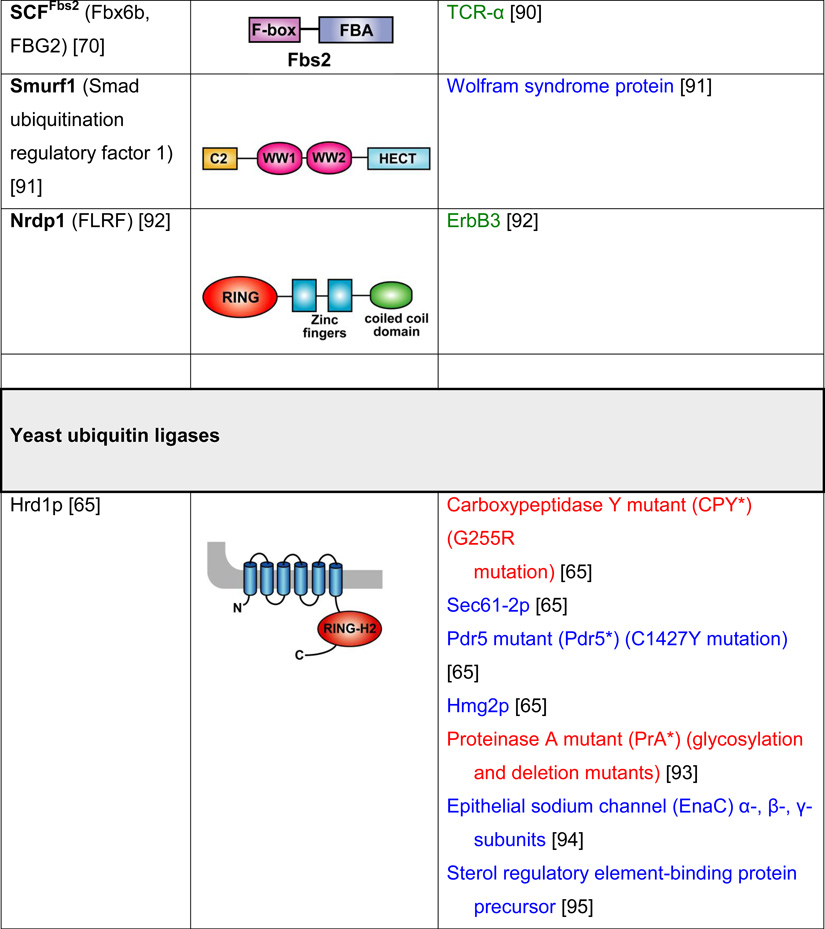
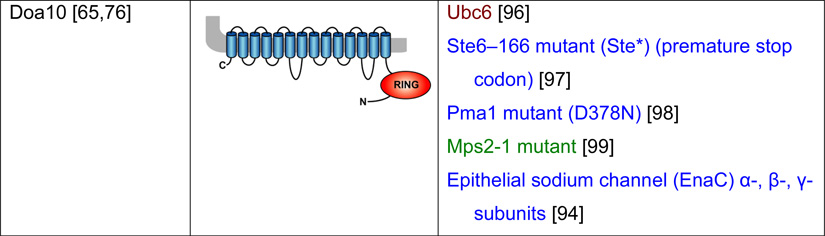
Table 2. Mammalian and yeast ubiquitin ligases involved in ER dislocation and corresponding in vivo substrates.
The ER membrane is represented by a shaded bar with the ER lumen in the upper right corner. Except for TEB4/Doa10, the indicated membrane topologies of polytopic ubiquitin ligases are predicted based on the sequence but not experimentally confirmed. Polytopic membrane substrates are in blue, single-pass membrane substrates are in green, ER luminal substrates are in red, and tail-anchored substrates are indicated in brown color. RING (really interesting new gene); PA (protease-associated domain); TPR (tetratricopeptide repeat domain); IBR (in-between RING domain); FBA (F-box-associated domain); HECT (homologous to the E6-AP carboxyl terminus).
 |
 |
 |
 |
 |
 |
